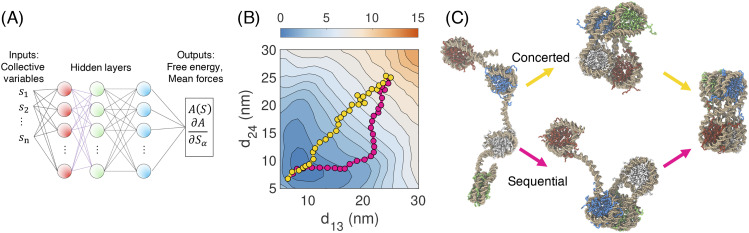
FIG. 9.
Stability and folding pathways of the tetra-nucleosome. (a) Illustration of the neural network approach for parameterizing high-dimensional free energy surfaces from mean forces. The neural network takes the six internucleosomal distances (S1, S2, …, Sn) as an input to compute the corresponding free energy [A(S)] and mean forces (). (b) Projection of the six-dimensional free energy profile to the distance between 1 and 3 (d13) and 2 and 4 (d24) nucleosomes. The sequential (pink) and concerted (yellow) pathway for tetra-nucleosome folding are shown on top of the free energy profile with energy unit kcal/mol. (c) Example tetra-nucleosome configurations along the two folding pathways. The DNA molecule is shown in gold, and the histone octamers are shown in green, white, blue, and red.