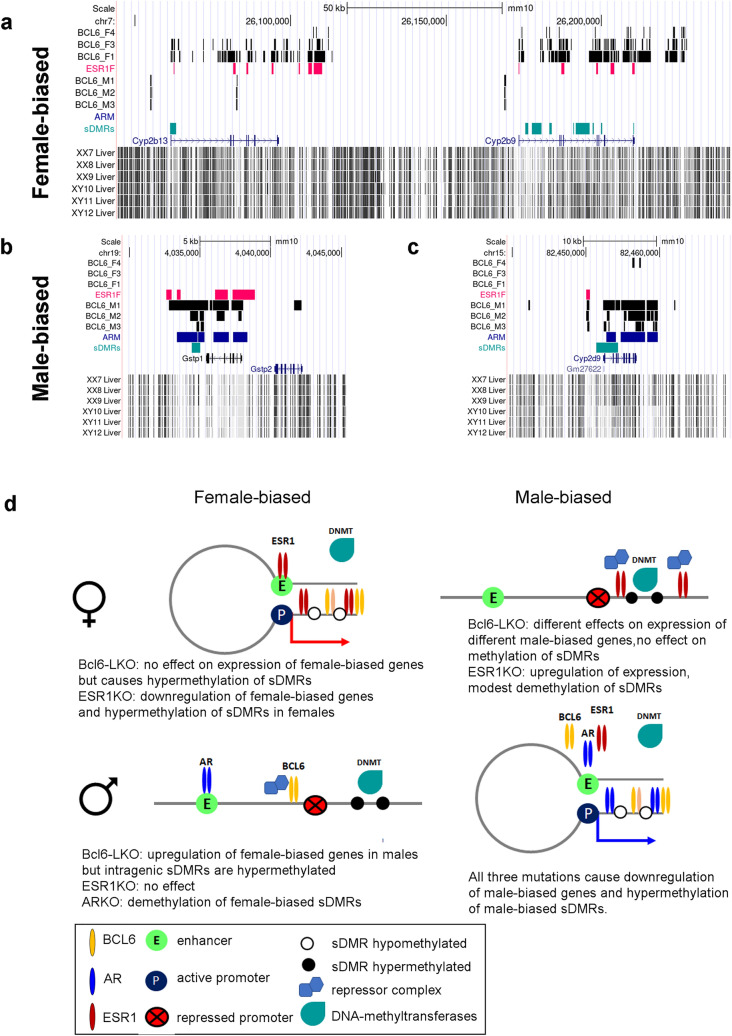
Figure 7.
Co-localization of AR, ESR1, and BCL6 enrichment with sDMRs and proposed working hypothesis. (a–c) Examples of overlapping sDMRs, sDEGs, BCL6, AR, or ESR1 enrichment; at female-biased sDMRs (a); and male-biased sDMRs (b and c). All tracks are shown in the context of the UCSC genome browser (mm10). BCL6-F1, BCL6-F3, and BCL6-F4 tracks show ChIP-seq peaks for BCL6 in the female liver (replicates 1, 3, and 4, respectively). BCL6-M1, BCL6-M2, and BCL6-M3 show ChIP-seq peaks for BCL6 in the male liver (replicates 1, 2, and 3, respectively)12. The ESR1F track shows ChIP-seq peaks for ESR1 in the female liver, and the ARM track shows ChIP-seq peaks for AR in the male liver30. The “sDMRs” track shows positions of sDMRs from the XX.F versus XY.M comparison based on WGBS data from6. The XX7, XX8, XX9, XY10, XY11, and XY12 liver tracks show positions of CG sites and the intensity of the tick reflects methylation level (WGBS data from6), in females (XX) and males (XY). (d) Schematic representation of the working hypothesis.