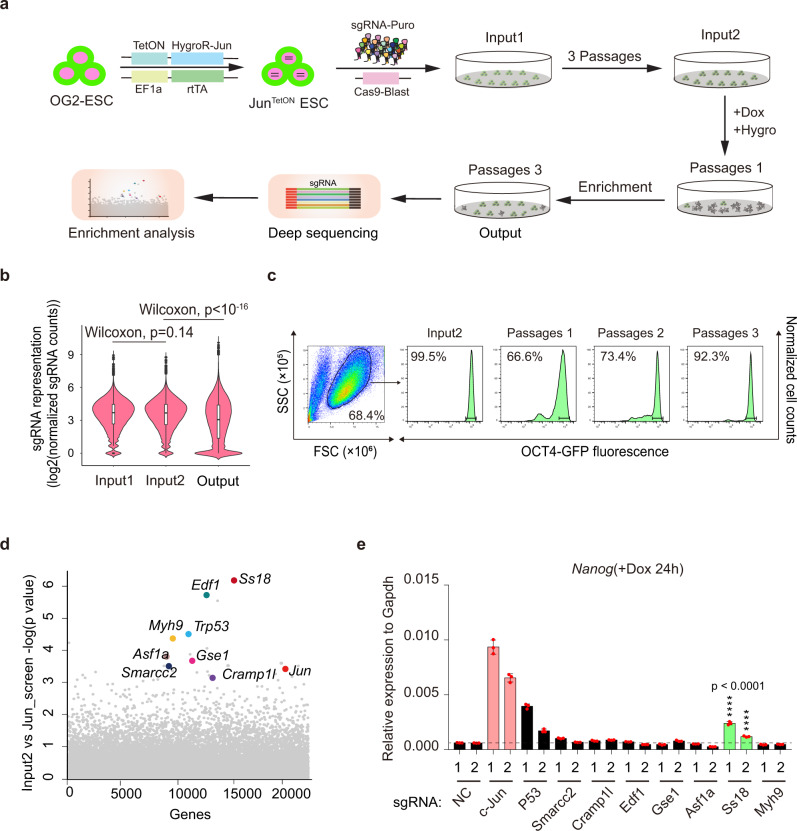
Fig. 1. Genome-wide screen for factors regulating PST.
a The workflow for GeCKO screening strategy by HygroR-JunTetON OG2ES. TetON cell line was successively infected with viruses expressing Cas9 and sgRNA. Dox and hygromycin were added after three cell passages. Dox and hygromycin were removed in 24 h and 48 h, respectively. Cells that survived after three passages were collected and analyzed. Hygro, hygromycin; HygroR, hygromycin resistance gene; GeCKO: Genome-wide CRISPR-Cas9 knockout. b Violin plot showing the distribution of sgRNA frequencies at different stages of screen. The distribution of sgRNA library was significantly polarized after the screening (output) compared to input 1 and input 2. Two-sided Wilcoxon test adjusted for multiple comparisons. n = 65959 sgRNAs. Input 1, Minimum 0, 25%Percentile 2.672, Median 3.693, 75%Percentile 4.452, Maximum 8.934; Input 2, Minimum 0, 25%Percentile 2.611, Median 3.672, 75%Percentile 4.456, Maximum 8.764; Output, Minimum 0, 25%Percentile 1.366, Median 3.064, 75%Percentile 4.391, Maximum 10.07. Two independent experiments. c Distribution of OG2-GFP signaling at different passages by FACS during this screening. d Identification of top candidate genes using the MAGeCK43 algorithm. e Validation of the hits in (d) by corresponding sgRNAs within GeCKO library during c-JUN induced PST. Data are mean ± s.d., two-sided, unpaired t test; n = 3 independent experiments, ****p < 0.0001.