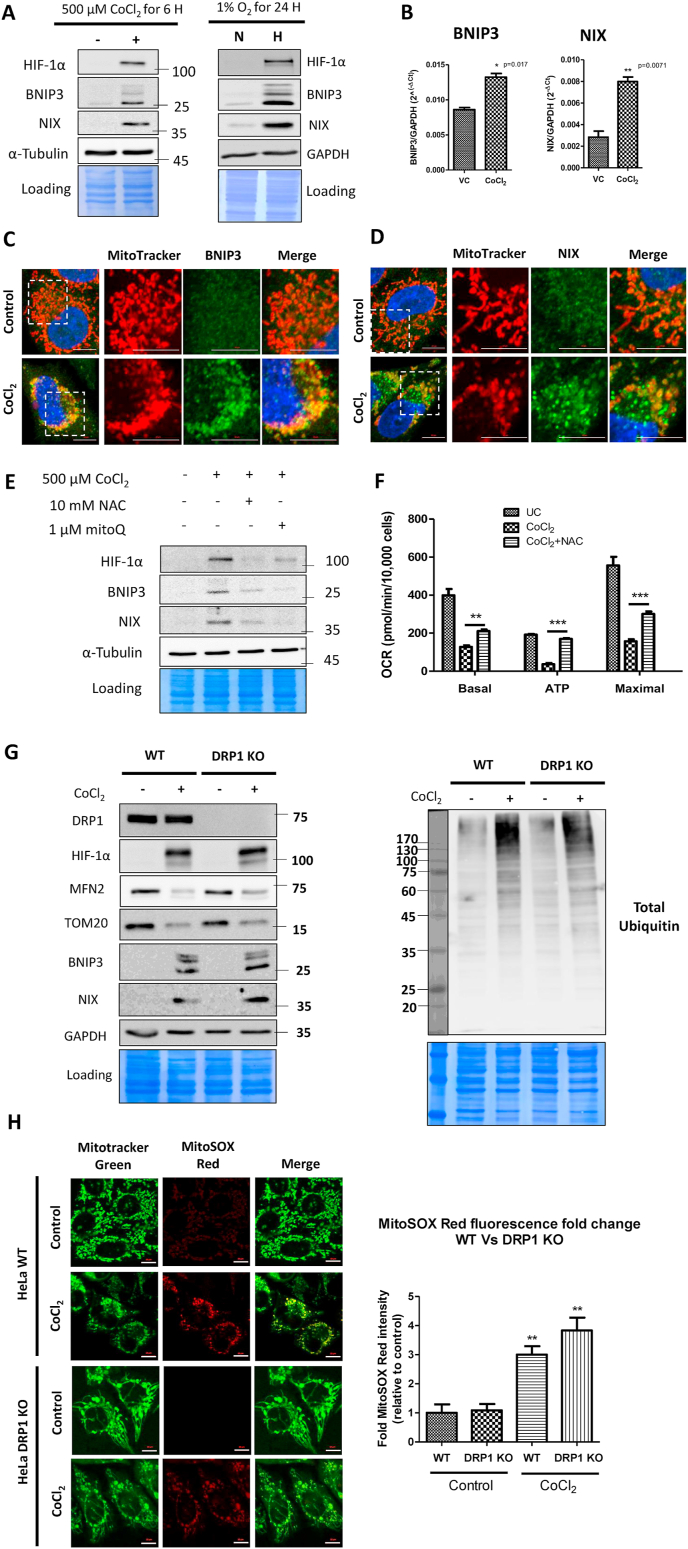
Fig. 4.
BNIP3 and NIX proteins drive mitophagy of oxidatively damaged mitochondria. A. Whole cell extracts of CoCl2 (500 μM CoCl2 for 6 h) and hypoxia-treated (1% O2 for 24 h) WT HeLa cells were analyzed for the expression of HIF-1α, BNIP3, NIX, GAPDH and Tubulin proteins by immunoblotting. B. The expression of BNIP3 and NIX transcripts in response to CoCl2 treatment (500 μM CoCl2 for 6 h) was analyzed by SYBR Green real time PCR relative to GAPDH and expressed as 2−ΔCt. WT HeLa cells treated with 500 μM CoCl2 plus 25 μM Z-VAD-FMK for 16 h were stained with Mitotracker Red followed by immunostaining with BNIP3 (C) and NIX (D). E. Following the indicated treatments for 24 h, the whole cell extracts of WT HeLa cells were analyzed for HIF-1α, BNIP3, NIX and Tubulin proteins by western blotting. F. The mitochondrial bioenergetics were studied by Seahorse flux analyzer. G. The whole cell extracts of control and CoCl2-treated WT and DRP1 KO HeLa cells were analyzed for DRP1, HIF-1α, MFN2, TOM20, BNIP3, NIX, total ubiquitin and GAPDH proteins (loading control) by immunoblotting. H. The control and CoCl2-treated WT and DRP1 KO HeLa cells were stained with MitoSOX red to evaluate the extent of ROS generated in response to CoCl2-induced oxidative stress and the MitoSOX red staining intensity was expressed as fold change relative to the control. Scale bar for microscopy images: 10 μm. Data represented as Mean + SEM of three experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)