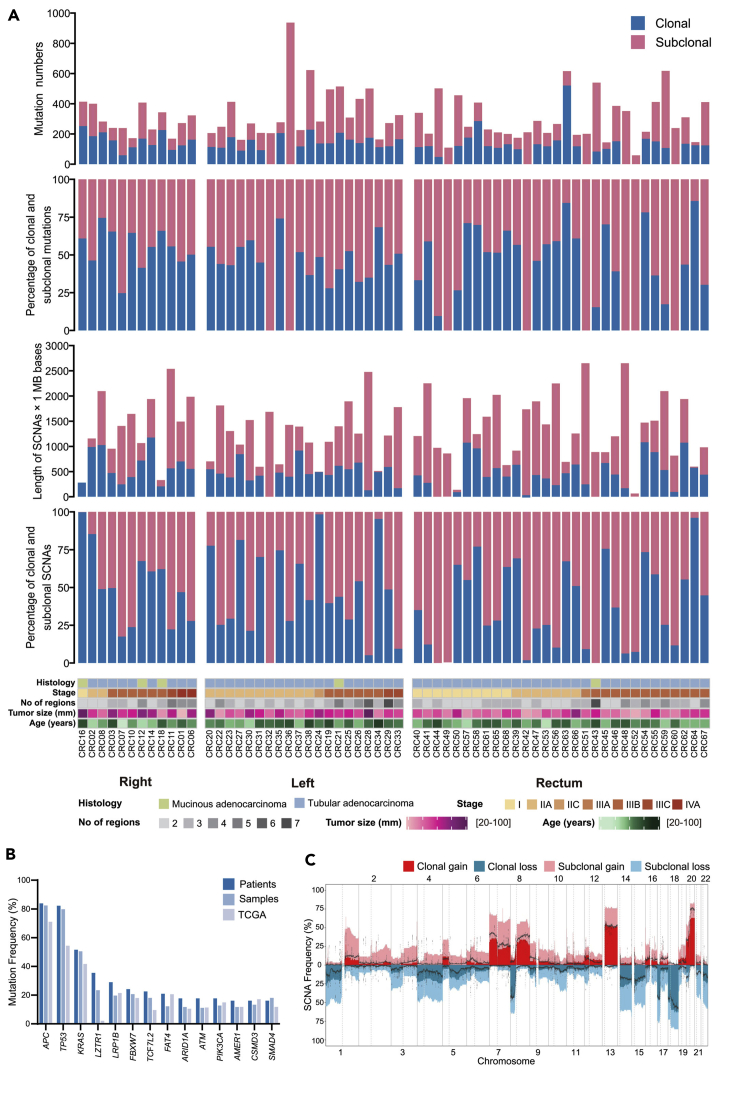
Figure 1.
Overview of genomic heterogeneity in CRC tumors
(A) Heterogeneity of mutations and somatic copy-number alterations (SCNAs). Tumors were sorted by location and stage. (1) Number of all SNV and INDEL mutations (including coding and noncoding mutations) in CRC tumors. (2) The percentages of clonal mutations in CRC tumors. (3) Quantification of SCNAs in CRC tumors. (4) The percentages of clonal SCNAs in CRC tumors. (5) Demographic and clinical characteristics of the 62 patients with CRC in this study (divided by histology; stage; number of regions; tumor size; age and tumor location).
(B) Mutation frequency of driver genes (driver mutations occurred in not less than 10 patients) and comparison with TCGA data. (C) Frequency of SCNAs in CRC tumors. The dotted lines were frequency of SCNAs in TCGA CRC samples.