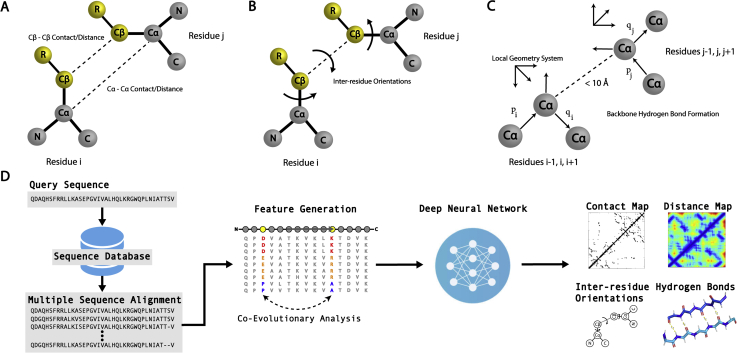
Figure 4.
Interresidue spatial restraints that are often used to assist protein 3D structure assembly simulations. The protein backbone atoms include the N, Cα, and C atoms, while the side chains include the Cβ atoms, with the exception of glycine, as well as the R groups, which distinguish the different amino acid residues. A, Cα/Cβ contacts and distances; B, interresidue torsion angles; C, hydrogen bond networks. Here, the backbone hydrogen bonds are represented using a Cα-based model, where three consecutive Cα atoms form a local coordinate system, from which various vectors and their orientations represent regular hydrogen bonding patterns observed in native proteins. D, typical pipeline for spatial restraint prediction. Starting from the amino acid sequence of a target protein, homologous protein sequences are collected from sequence databases and compiled to form a multiple sequence alignment (MSA). For the MSA, coevolutionary relationships are deduced and fed into a deep neural network, which may output the predicted contact/distance maps, interresidue orientations, and hydrogen bond networks.