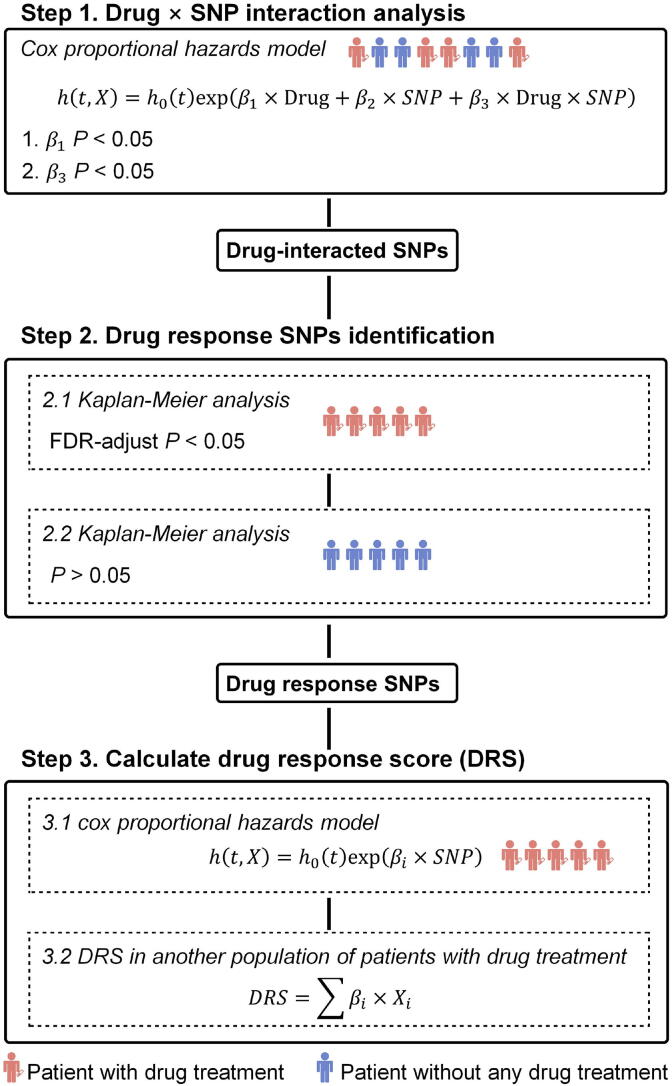
Fig. 1.
An overview of the approach. Step 1: We performed SNP × drug interaction analysis in all patients (including patients with drug treatment and patients without any drug treatment). SNPs with significant drug term (P < 0.05 for) and significant interaction term (P < 0.05 for) were remained. Step 2: For SNPs obtained from the first step, we performed Kaplan-Meier (KM) analysis in subjects with different genotypes to select SNPs associated with drug response in patients with drug treatment. KM analysis in patients without any drug treatment was also performed to remove the SNPs associated with survival but this association was not related to drug treatment. Step 3: We performed univariate cox proportional hazards analysis for each drug response SNPs get from the first two steps. The coefficients for all SNPs was used to calculate drug response score (DRS) in another population to test the performance of drug response prediction.