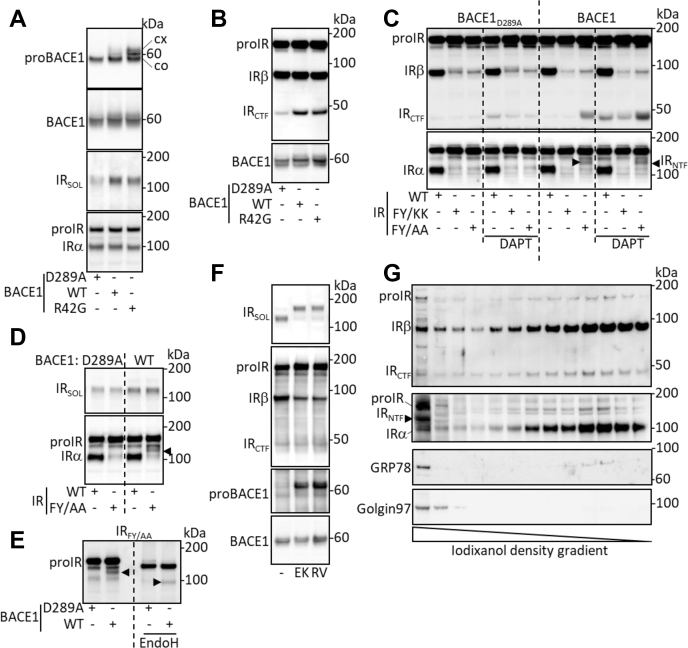
Figure 2.
proBACE1 cleaves proIR in the early secretory pathway. Wildtype (WT), inactive (D289A), or PC-resistant (R42G) forms of BACE1 were overexpressed with IR or its indicated mutated forms. A, proBACE1, total BACE1, and IRα subunit were detected in cell lysates; IRSOL was detected in conditioned media. B, cells were treated with DAPT (10 μM, 17 h), then IRβ subunit and BACE1 were detected in cell lysates. C, cells were treated with DAPT, then IRβ and IRα subunits were detected in cell lysates. D, IRα subunit and IRSOL were detected in cell lysates and in conditioned media, respectively. E, cell lysates were treated with EndoH, and IRα subunit was detected. F, BACE1 and IR-expressing cells were treated with Dec-RVKR-CMK (RV; 20 μM, 17 h) or transfected with EK4 expression vector (EK) to inhibit PC, then IRSOL was detected in the conditioned media and IRβ subunit, proBACE1, and BACE1 were detected in cell lysates. G, distribution of IR, IRCTF, and IRNTF in subcellular membrane vesicles of HEPG2 cells fractionated by iodixanol density gradient. Golgi apparatus- and ER-rich fractions are identified by the detection of Golgin 97 and GRP78, respectively. Positions of IR precursor (proIR), IR C-terminal fragment (IRCTF), IR N-terminal fragment (IRNTF, black arrowhead), and core glycosylated (co) and complex N-glycosylated (cx) proBACE1 are indicated.