Figure 3.
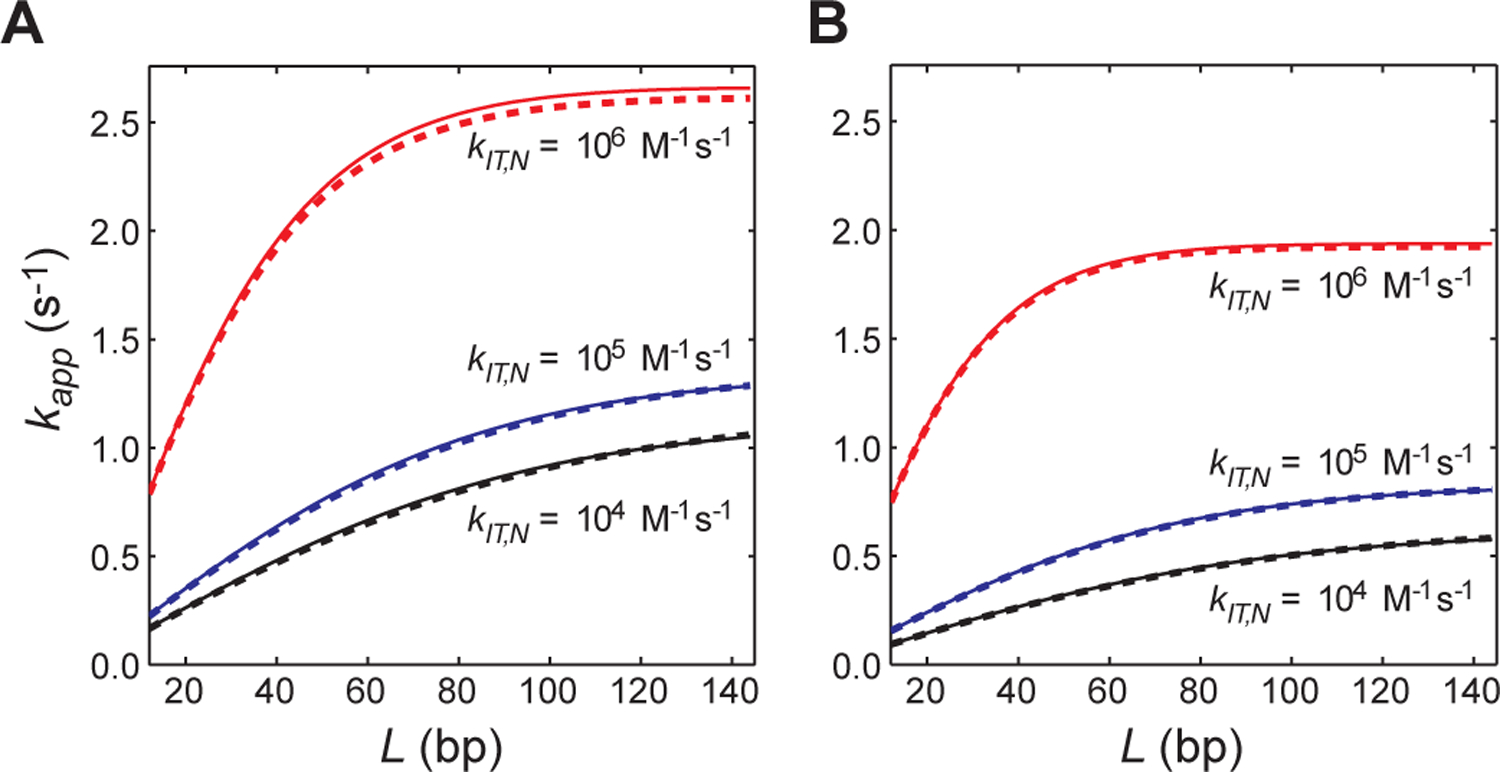
Validation of the VK model-based analytical forms of kapp for systems involving competitor DNA and intersegment transfer (Eq. 18). (A) Rate constant kapp for target association as a function of the total number of sites L. A single orientation per site (i.e., ϕ = 1) was assumed. The sliding rate constant ksl,N = 105 s−1 was used. The other conditions are exactly the same as those for Figure 2A except that intersegment transfer is taken into consideration. The rate constants for intersegment transfer are indicated. (B) Rate constant kapp for target association as a function of the total number of sites L under conditions identical to those for the panel A, except that 2 orientations per site are assumed (i.e., ϕ = 2). The kinetic and thermodynamic parameters are defined for each orientation. For both panels, the dashed lines were obtained from the ODE-based numerical kinetics simulations, whereas the solid lines were obtained with Eq. 18. Details of the ODE-based simulations are given in the Supplemental Information.
