FIGURE 1.
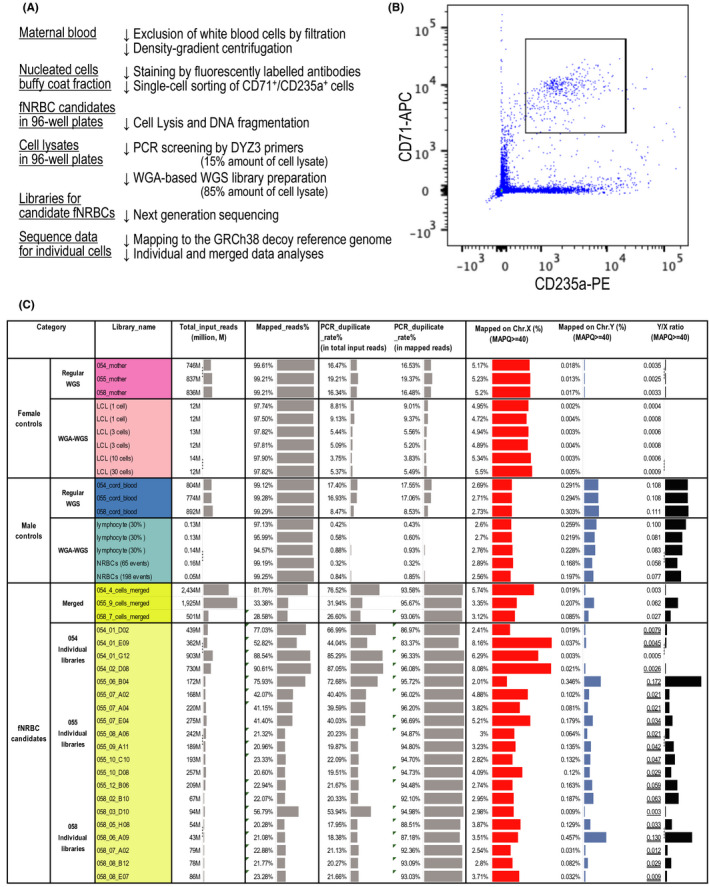
A. A workflow to isolate and to characterize single fNRBCs as adopted in this study. B. A FACS dot plot of nucleated cells isolated from blood of a pregnant woman (Case 058). Boxed CD71 and CD235a double‐positive cells were sorted. C. Mapping metrics of the sequencing data of the regular WGS and WGA‐based WGS libraries. Female control libraries include three regular WGS libraries prepared from 600 ng genomic DNA of maternal blood and six WGA–WGS libraries prepared from cells of a lymphoblastoid cell line (LCL). Male control libraries include three regular WGS libraries prepared from 600 ng genomic DNA of cord blood of the newborn, three WGA–WGS libraries prepared from 30% of the amount of the cell lysate of a single lymphocyte, and the WGA–WGS libraries prepared from pools of NRBCs isolated from cord blood (65 and 196 FACS events). Sequencing data of the WGA–WGS libraries prepared for fNRBC candidates were analyzed individually and all together for each case (“merged”). The Y/X ratio value of an fNRBC candidate was double undermined or single undermined, when it surpassed a threshold for the presence of Y chromosome, that is, the average Y/X ratio plus 3 SD value of the female control regular WGS libraries (0.0044), or that of the female control WGA–WGS libraries (0.0012)
