Figure 3.
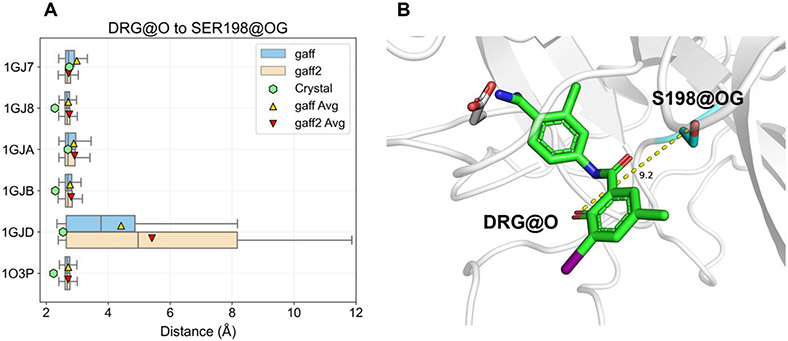
Relieving steric clash between the ligand phenol and Ser-198. (A) The distance between the ligand phenol oxygen and Ser-198 hydroxyl oxygen is recorded over the last 10 ns of equilibration to analyze sampled conformations and compared to the distance observed in the crystal structures. The trend observed is identical for both GAFF and GAFF2 force fields, all ligands except 1GJD twist away due to repulsive steric interactions but remain in hydrogen bonding range. 1GJD samples broad distances, indicating the initial hydrogen bond is detached. (B) Sample frame from the 1GJD simulation illustrates that the phenol hydroxyl rotates outward away from the protein, and the starting hydrogen bond is replaced with one between the peptide bond-like carbonyl and Ser-198. The inhibitor is colored green and labeled DRG.