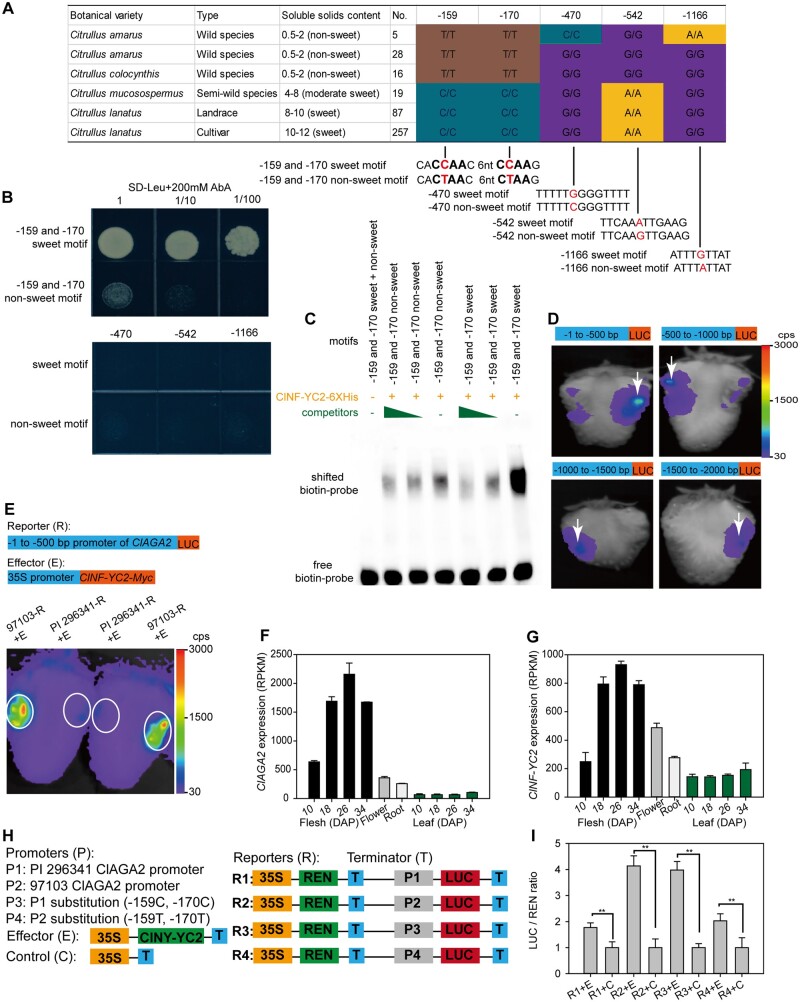
Figure 4.
ClNF-YC2 regulates ClAGA2. A, Haplotype of ClAGA2 gene promoter. Homozygous genotype “A,” “T,” “C,” and “G” was shown as yellow, brown, cyan, and purple. B, Y1H experiment coexpressing transcription factor ClNF-YC2 with sweet or nonsweet motifs in ClAGA2 promoter. C, EMSAs of ClNY-YC2 fusion protein binding to the cis-element in ClAGA2 from sweet 97103 and nonsweet PI 296341. D, LUC activity in strawberry fruits expressing the four 5′-truncated fragments of the ClAGA2 gene promoter reporter constructs. The white arrows indicate injection sites of Agrobacterium. Color in scale represents the fluorescence intensity in counts per second (cps). E, LUC assay of strawberry fruits cotransformed with ClNY-YC2 as the effector and the −500 bp promoter region of ClAGA2. Strong LUC activity was detected with the promoter of the sweet (97103), while no LUC activity was detected with PI 296341-pClAGA2::LUC construction. Two representative results of five independent experiments are shown. The white circles indicate sites for Agrobacterium-mediated infiltration. Scale shows the fluorescence intensity from 30 to 3000 cps. F, Relative expression levels of ClAGA2 in sugar loading tissue (leaf) and unloading tissues (fruit, flower, and root) during plant development. Error bars, sd values for n = 3 independent biological replicates. G, RNA-seq analysis of expression levels of ClNY-YC2 in sugar loading and unloading tissues at different stages. H, Diagram of the various constructs with the –1,512 bp promoter used for dual-LUC reporter assays. I, The reporter and effector vectors were cotransformed into watermelon fruit protoplasts. LUC and REN (Renilla) LUC activities were assayed, and the activation of ClNY-YC2 to the reporters was shown by the ratios of LUC to REN. The LUC/REN ratios of the empty vector cotransformed with reporters were set as 1 for controls. Each value represents the mean ± sd of five biological replicates. **t-test significant at P < 0.01.