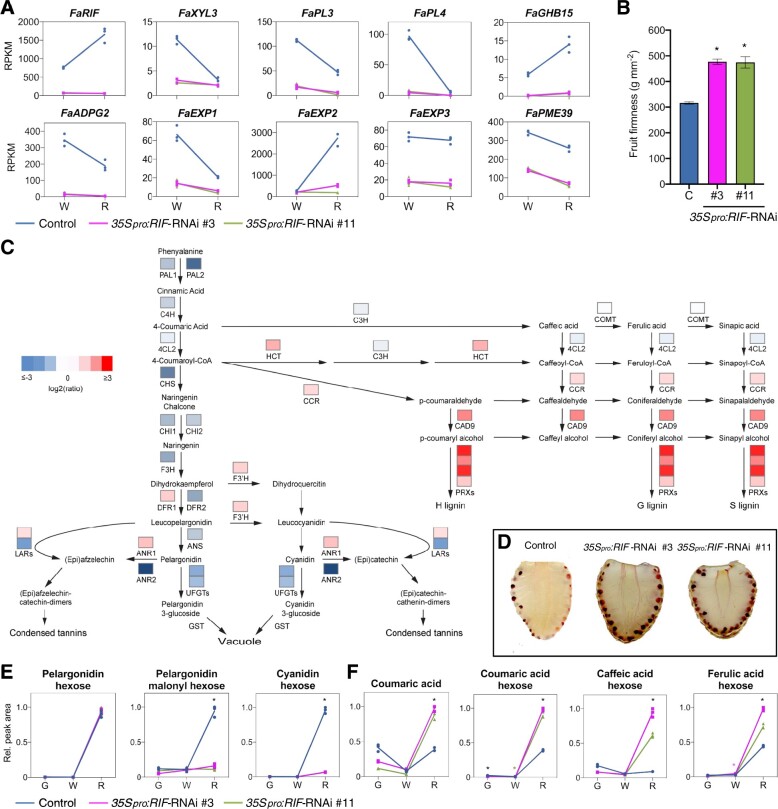
Figure 3.
FaRIF controls the expression of genes involved in cell-wall degradation and the phenylpropanoid pathway, controlling fruit firmness, and anthocyanin and lignin levels. A, Expression of FaRIF and genes involved in cell-wall degradation and modification in control and 35Spro:RIF-RNAi white (W) and red (R) receptacles. B, Fruit firmness measurement in control (C) and 35Spro:RIF-RNAi ripe receptacles. Data are means ± se of 10 biological replicates analyzed by Student’s t-test (*P < 0.001). C, Phenylpropanoid, flavonoid, and lignin biosynthetic pathways. Colors denote the average of the log2 of the 35Spro:RIF-RNAi/control expression ratio in both transgenic lines at the red stage for the respective genes. Red and blue show up- and downregulation, respectively, in both silenced lines. D, Lignin staining in ripe fruit sections from control and 35Spro:RIF-RNAi plants using phloroglucinol. Photographs were taken at the same distance. E, F, Changes in relative contents of anthocyanins (E) and hydroxycinnamic acid derivatives (F) in green (G), white (W), and red (R) receptacles of control and 35Spro:RIF-RNAi receptacles. Data in (E) and (F) were analyzed by Student’s t test (*P < 0.0005). Black asterisks indicate P < 0.0005 for both RNAi lines compared with the control. Colored asterisks denote P < 0.0005 for one of the RNAi lines compared with the control.