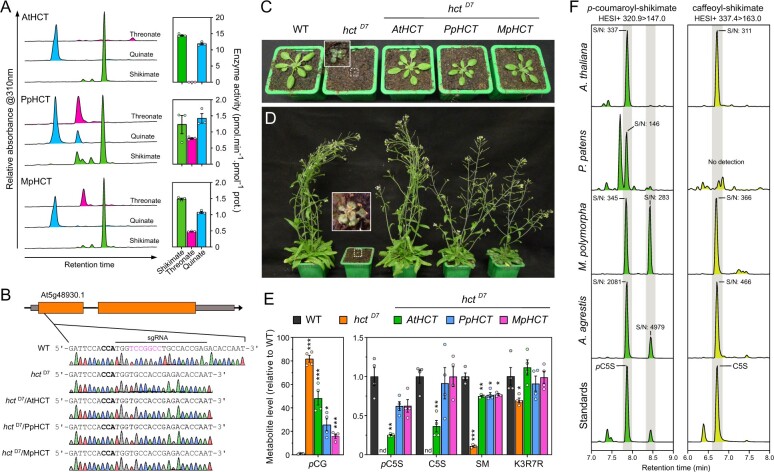
Figure 6.
Evolutionary conservation of HCT function in embryophytes. A, AtHCT, PpHCT, and MpHCT acyl acceptor permissiveness was investigated using threonate, quinate, or shikimate, and p-coumaroyl-CoA as an acyl donor in in vitro end-point assays. Representative HPLC-UV chromatograms (left) and corresponding HCT activity (right) are shown for the different acyl acceptors. Results are the mean ± SEM of three independent enzyme assays. Note that the results for PpHCT are the same as those reported in Figure 3, C. B, Schematic representation of the AtHCT locus and sequence of the CRISPR/Cas9 target site. Gray box, UTR; black line, intron; orange box, exon. The protospacer adjacent motif (NGG) is highlighted in bold. Sanger sequencing chromatograms of wild-type (WT) and the homozygous hctD7 mutant transformed, or not, with AtHCT, PpHCT, or MpHCT show the seven-nucleotide (in pink) deletion in the HCT gene of hctD7 plants. C, D, Phenotypes of 3-week-old (C) and 60-day-old (D) A. thaliana WT and the hctD7 null mutant transformed, or not, with AtHCT, PpHCT, or MpHCT genes. E, Relative levels of the phenolic esters p-coumaroyl-glucose (pCG), p-coumaroyl-5-O-shikimate (pC5S), caffeoyl-5-O-shikimate (C5S) and sinapoyl-malate (SM), and of the flavonol kaempferol 3-O-rhamnoside 7-O-rhamnoside (K3R7R) in the rosettes of 3-week-old plants. Results are the means ± SEM of four independent biological replicates. WT versus mutant t test adjusted P-value: *P<0.05; **P<0.01; ***P<0.001. F, Representative UHPLC-MS/MS chromatograms reporting the search for p-coumaroyl-5-O-shikimate and caffeoyl-5-O-shikimate in the bryophytes P. patens, M. polymorpha and A. agrestis, and the angiosperm A. thaliana. Additional positional isomers of p-coumaroyl-shikimate may occur in M. polymorpha and A. agrestis. S/N, signal-to-noise ratio. Gray shaded regions highlight relevant elution time windows.