Figure 1.
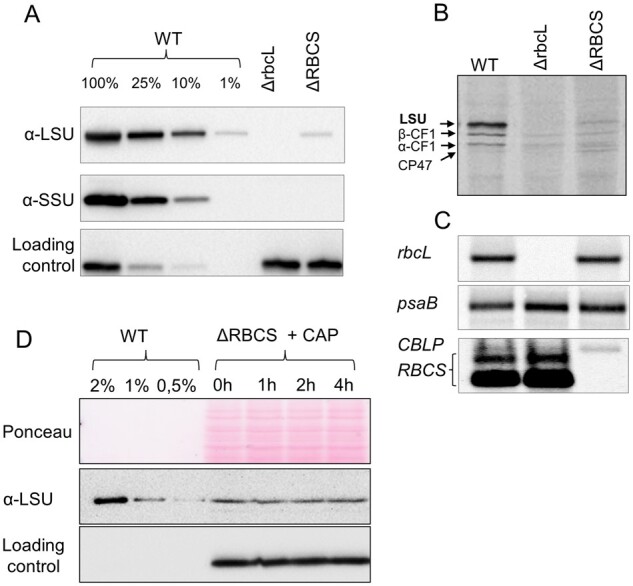
LSU accumulation, synthesis rate, and stability in the absence of its assembly partner. (A) Immunoblot showing protein accumulation of Rubisco subunits in the ΔRBCS strain, using an antibody directed against whole Rubisco holoenzyme. PsaD accumulation, revealed with a specific antibody, is shown as a loading control. (B) 14C pulse labeling experiment showing the synthesis rate of LSU in the ΔRBCS strain as compared to the WT in upper panel (positions of LSU as well as ATPase α and β subunits and PSII CP43 subunit are indicated by arrows). (C) mRNA accumulation in the same strains as in B, as probed by hybridization with rbcL and RBCS probes, and psaB and CBLP probes used as loading controls. In both panels, the ΔrbcL strain exhibiting a deletion of the rbcL gene is used as a negative control. (D) Unassembled LSU stability assayed by immunochase over 4 h after chloroplast synthesis arrest by chloramphenicol (CAP) addition. LSU is detected with the anti-Rubisco antibody, cyt f is used as a loading control.
