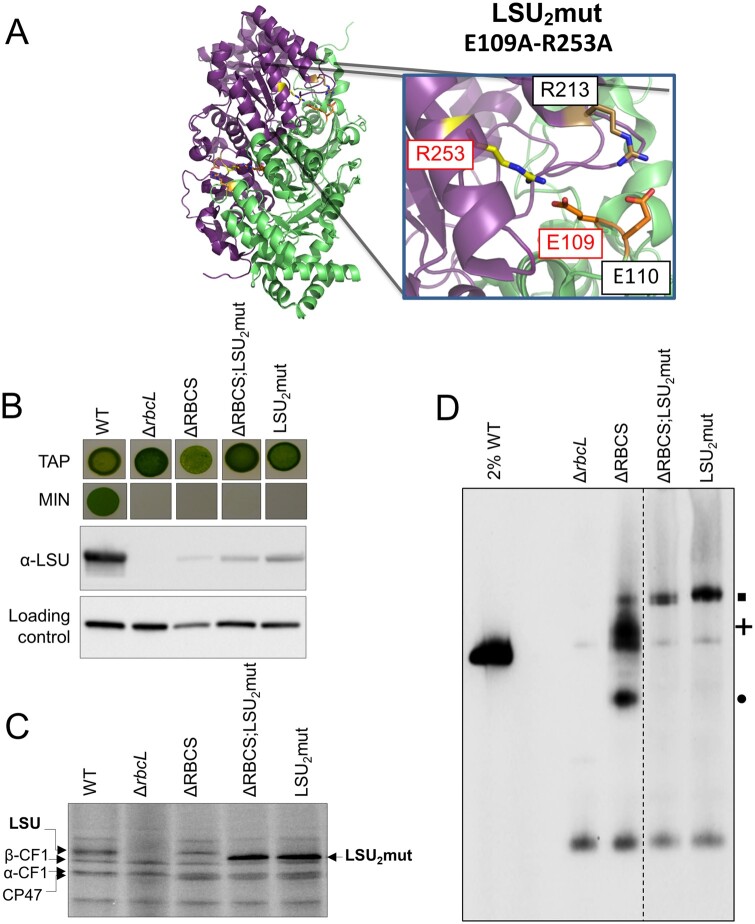
Figure 8.
LSU2 mutations alter LSU accumulation and CES regulation. (A) Close-up of the mutated residues in LSU2mut strain in LSU structure. C. reinhardtii LSU dimer structure is shown in cartoon, as extracted from Rubisco structure (PDB: 1IR2). The two LSU subunits forming the dimer are represented in green and magenta. Subunits are maintained by two inter-subunits salt bridges between E109 and R253, and E110 and R213 residues. Residues mutated in LSU2mut (E109A and R253A) are highlighted in red. The figure was generated using the PyMol program (Schrödinger-LLC). (B) Impairment in Rubisco accumulation is revealed by the absence of phototrophic growth in the LSU2mut and ΔRBCS;LSU2mut strains as probed by spot tests on acetate-free minimal media (MIN). Growth on TAP is shown as a control. The corresponding soluble LSU accumulation detected by immunoblot is shown together with PsaD accumulation as loading control. (C) LSU synthesis rate in LSU2mut and ΔRBCS;LSU2mut measured by short 14C pulse labeling experiment and compared to WT. Note that in the 12–18% acrylamide-8M urea gel system, the mutated LSU undergoes a change in its migration pattern compared to native LSU. (D) Immunoblot with the Rubisco antibody after CN-PAGE analysis of soluble protein fractions of WT (note the dilution), ΔrbcL, ΔRBCS, LSU2mut and ΔRBCS;LSU2mut strains. A dashed line marks the position where two irrelevant lanes were removed. The position of the LSU-complexes observed in ΔRBCS is indicated at the right using the same symbols as in Figure 5 (square, cross, and circle).