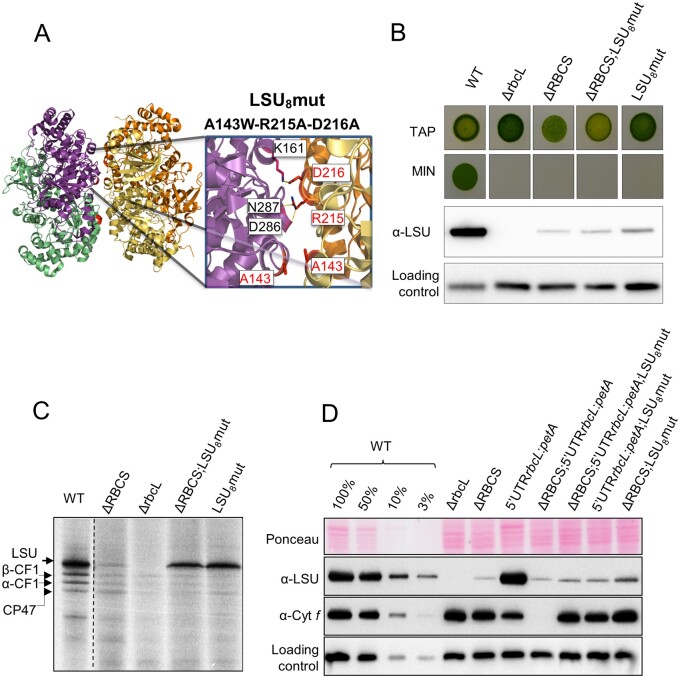
Figure 9.
Disruption of LSU oligomerization alters LSU CES regulation. (A) Close-up of the mutated residues in the LSU8mut strain in LSU structure. Two LSU dimers facing each other are shown, as extracted from C. reinhardtii Rubisco structure (PDB: 1IR2). LSU subunits from the first and second depicted dimers are shown respectively in green and magenta, and in orange and yellow. The dimer to dimer interaction is stabilized by hydrogen bonds between the R215 and D286-N287 residues, and by a salt bridge involving the D216 and K161 residues, which are represented on the cartoon. The distance between the two dimers is the shortest at the A143 residues facing each other. Residues mutated in LSU8mut (ARD) are highlighted in red. The figure was generated using the PyMol program (Schrödinger-LLC). (B) Impairment in Rubisco accumulation is revealed by the absence of phototrophic growth in the LSU8mut and ΔRBCS; LSU8mut strains as probed by spot tests on acetate-free minimal media (MIN). Growth on TAP is provided as a control. Control strains WT, ΔrbcL, and ΔRBCS come from the same cultures as the ones used in Figure 7A. The corresponding soluble LSU accumulation detected by immunoblot is shown. (C) LSU synthesis rate in LSU8mut and ΔRBCS;LSU8mut measured by short 14C pulse labeling experiment and compared to ΔRBCS and WT. The dashed line marks the position of two irrelevant lanes, which were removed. (D) Immunoblot showing LSU and cyt f accumulation levels in the wild-type (WT), ΔRBCS, ΔrbcL, LSU8mut, and ΔRBCS;LSU8mut strains and in those latter three genetic contexts combined with the 5′UTRrbcL:petA reporter gene background. PsaD accumulation is provided as a loading control.