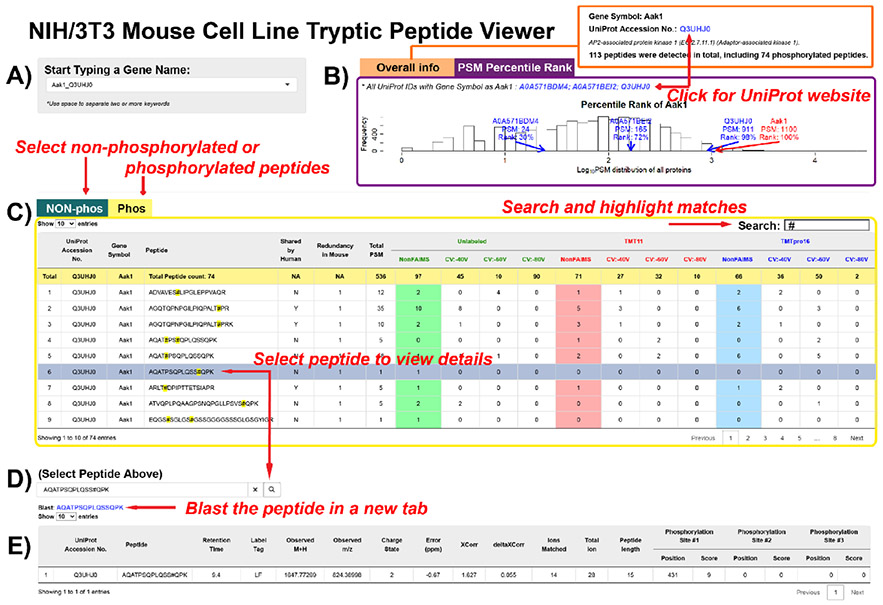
Figure 5.
The NIH/3T3 Mouse Cell Line Tryptic Peptide Viewer. (A) Proteins may be selected from a drop-down list by typing in key letters.. Gene symbols and UniProt accession numbers are listed in pairs. (B) Tab panels with overall information in one tab, and PSM information in the other tab. In the “Overall Info” tab, total peptide numbers are summarized. A link to the UniProt entry of the selected protein is provided. In the “PSM Percentile Rank” tab, a histogram showing the log10(PSM) distribution is presented. PSMs of the proteins with selected gene symbol and the percentile ranks of the log10(PSM) are labeled on the plot. (C) Nonphosphorylated peptides are listed in one tab, and phosphorylated peptides listed in another. The data table outputs all the peptides assigned to the selected protein and the peptide spectrum match (PSM) numbers under different analytical conditions. The search box on the right corner allows character searches, such as amino acid residues. After clicking on a row, the peptide sequence will be copied into (D), a peptide search box. Clicking the search button submits the search, and a blast hyperlink will appear below. (E) The peptide detail table displays the peptides with modifications (if any), peptide characteristics, and the phosphorylation site localization score (if applicable). The application is available at https://wren.hms.harvard.edu/OPapp/.