Fig 3. Suppression of MAP3K7 and CHD1 increases enzalutamide resistance and AR-v7 signaling.
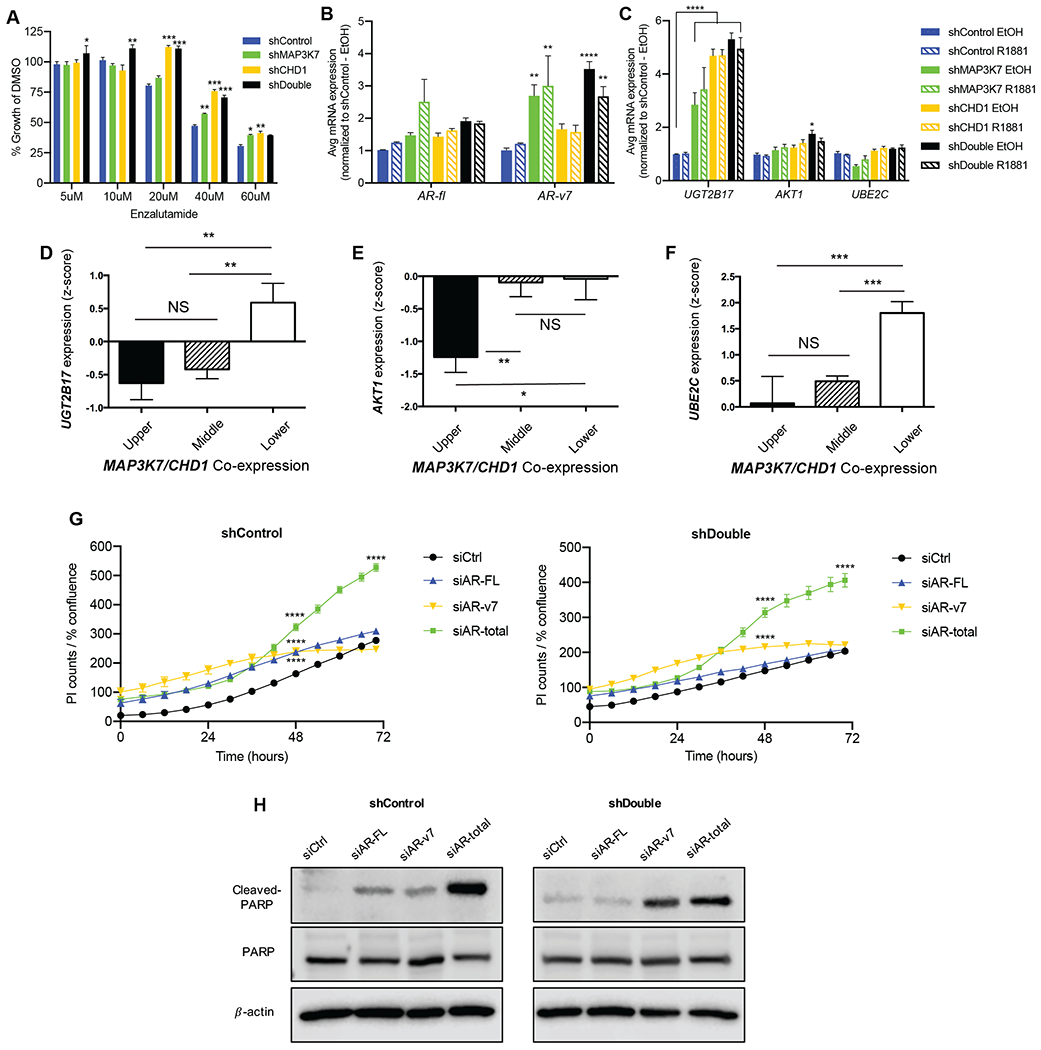
(A) Day 5 of growth assay for indicated LNCaP cells treated with 5-60 µM enzalutamide in complete medium, normalized to DMSO vehicle control (n=3; p-values indicate comparison to shControl at each concentration). (B) RT-qPCR of AR-FL and AR-v7 in indicated LNCaP cells ± 1 nM R1881 for 4 h. (C) RT-qPCR analysis of AR-v7 target genes UGT2B17, AKT1, and UBE2C in shControl and shDouble LNCaP cells ± EtOH or 1 nM R1881 for 4 h. For (B-C), Ct values normalized to B2M expression and then to shControl (n=3; p-values indicate comparison to shControl EtOH). (D) Correlation of UGT2B17, (E) AKT1, and (F) UBE2C mRNA expression with quartiles of MAP3K7 and CHD1 mRNA co-expression in the MSKCC dataset (25). (G) Propidium iodide (PI) counts normalized to cell confluence of shControl (left) and shDouble (right) LNCaP cells in castration conditions beginning 24 hours after transfection with indicated siRNAs (n=3; p-values indicate comparison to siCtrl). (H) Western blot of lysates from LNCaP cells 96 hours after transfection with indicated siRNAs showing cleaved PARP, total PARP, and the loading control β-actin. All data represent means ± SEM; unpaired two-tailed t-tests (D, E, F) and two-way ANOVA with Dunnett’s multiple comparisons test (A, B, C, G); *=p<0.05; **=p<0.01; ***=p<0.001; ****=p<0.0001.
