Figure 5. RNA-seq identifies cuprizone treatment-induced differential gene expression in hGPCs and oligodendroglia.
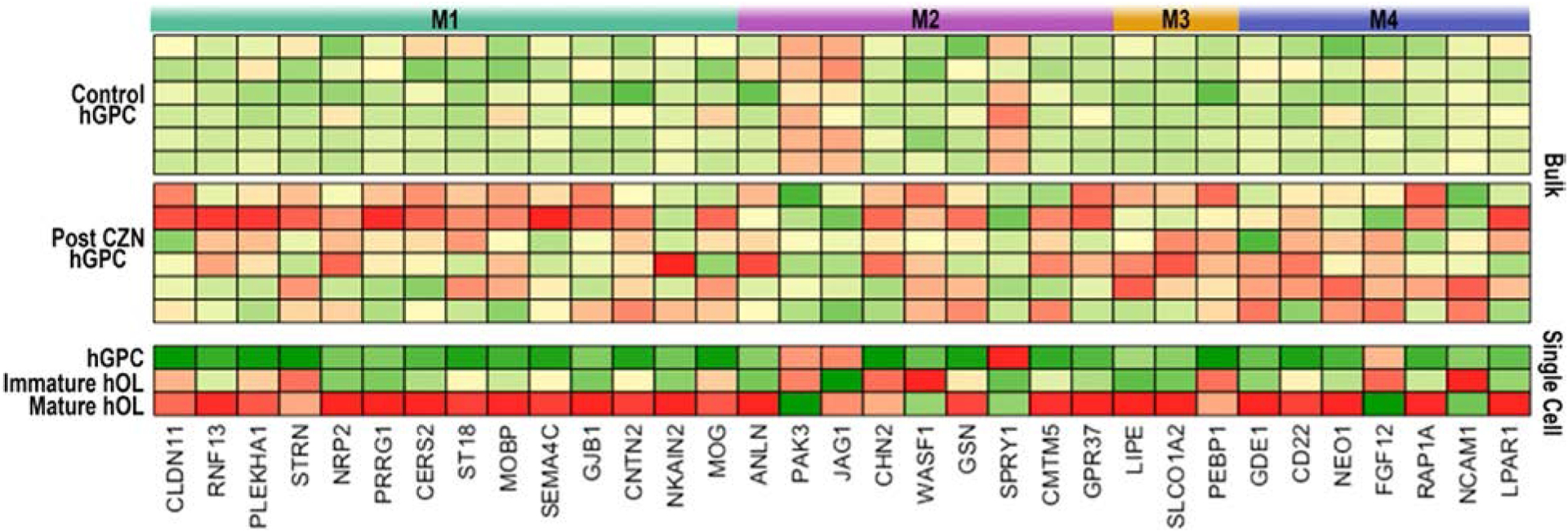
Bulk RNA-sequencing was done on hGPCs sorted via FACS from the corpus callosa of human glial chimeras. The mice were engrafted with fetal tissue-derived hGPCs at birth, given oral cuprizone (CZN) or a control diet for 12 wks beginning at 12 wks of age, and killed at 36 wks for expression profiling. A subset of differentially-expressed genes is shown, with comparison between the hGPC, immature OL, and mature OL pools identified in the human glial scRNA-seq data of Fig 4.
After gene ontology network analysis, major differentially-expressed genes were segmented into functional modules (M1–4). M1: myelination, TCF7L2 signaling; M2: Notch and TGFβ signaling, cell movement; M3: lipid and T3 transport, RXRA signaling; M4: iron and copper homeostasis, calcium signaling. Expression values are experiment-specific gene Z-Scores (red, high expression; green, low expression).
From (Windrem et al., 2020).
