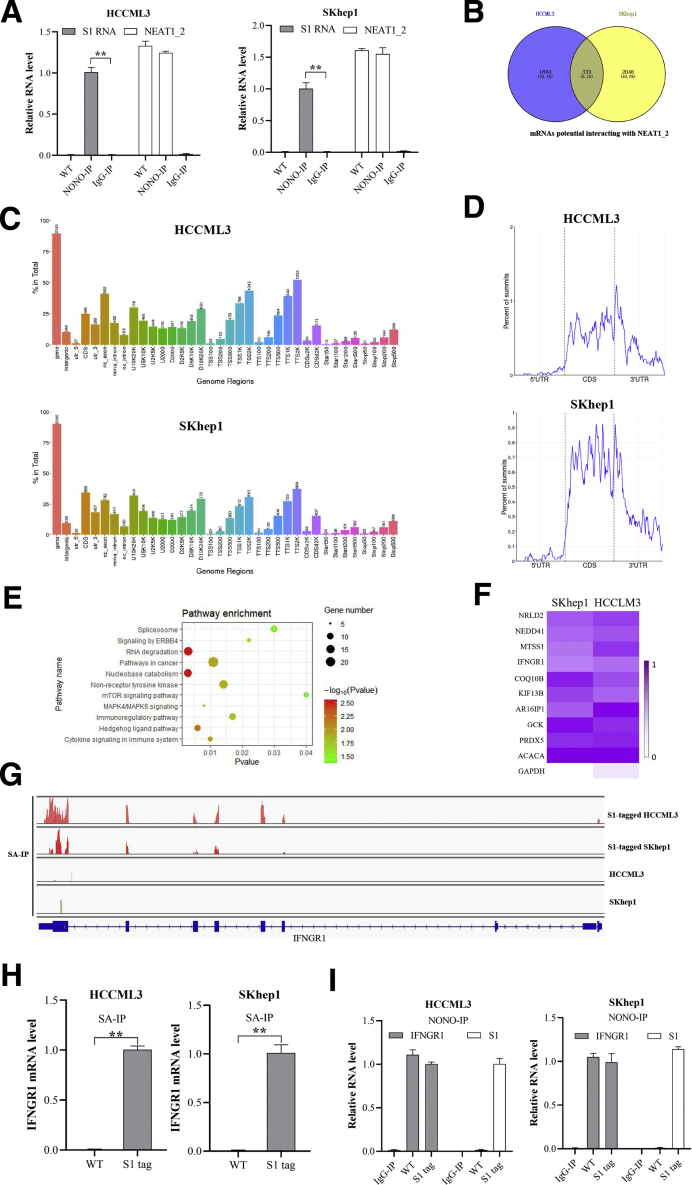
Figure 3.
IFNGR1 mRNA is abundant in paraspeckle of T-cell killing resistant HCC cells. (A) RIP analysis of S1 or NEAT1_2 relative expression in S1-tagged HCCML3 or SKhep1 cells with anti-NONO antibody. (B) Venn diagram of mRNAs potentially interacted with NEAT1_2 in S1-tagged HCCML3 or SKhep1 cells, determined by RIP-seq by precipitation with streptavidin. (C) Genome regions of transcripts potentially interacted with NEAT1_2 in S1-tagged HCCML3 or SKhep1 cells. (D) Binding regions of transcripts potentially interacted with NEAT1_2 in S1-tagged HCCML3 or SKhep1 cells. (E) Kyoto Encyclopedia of Genes and Genomes analysis of the overlapped genes in Figure 2B. (F) Heatmap of the top 10 genes of the overlapped genes. (G) Assay for transposase accessible chromatin with RNA-seq at the IFNGR1 locus. (H) RIP analysis of S1-tagged NEAT1_2 interacted with IFNGR1 mRNA with streptavidin. (I) RIP analysis of NONO interacted with IFNGR1 mRNA or S1-tagged NEAT1_2 with anti-NONO antibody. Data are represented as means ± SD (n = 3; ∗P < .05, ∗∗P < .01).