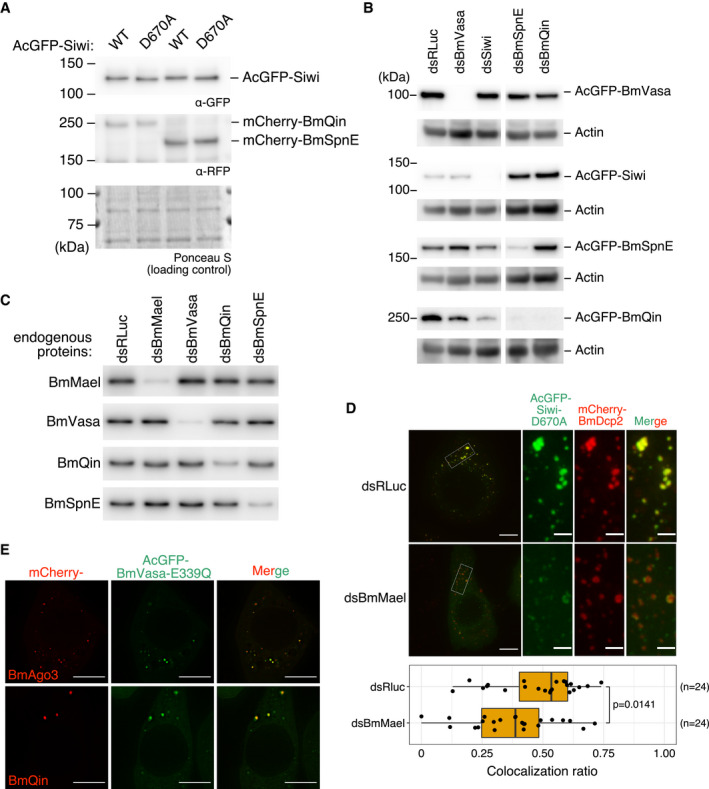
Western blotting of AcGFP‐Siwi, wild‐type (WT) or D670A, and the co‐expressed mCherry‐BmQin or mCherry‐BmSpnE. Expression of Siwi‐D670A did not affect the expression level of BmQin or BmSpnE.
Validation of dsRNA‐mediated knockdown of epitope‐tagged piRNA factors by Western blotting.
Validation of dsRNA‐mediated knockdown of endogenous piRNA factors by Western blotting.
Depletion of BmMael results in partial segregation of Siwi‐D670A foci from BmDcp2 foci. (Top‐left) Representative Z‐projections (Maximum intensity). Scale bar 8 μm. (Top‐right) Enlarged area from the white box. Scale bar 2 μm. (Bottom) Box plot showing that depletion of BmMael (dsBmMael, n = 24 cells) reduced colocalization ratio between Siwi and BmDcp2, compared with control (dsRLuc, n = 24 cells). Representative data from N = 3 independent experiments are shown. P‐value was calculated by asymptotic Wilcoxon rank sum test. Centre line, median; box limits, lower (Q1), and upper (Q3) quartiles; whiskers, 1.5 × interquartile range (IQR); points, outliers.
Colocalization of BmAgo3 and BmQin with BmVasa‐E339Q (ATPase mutant). Scale bar 10 μm.