Figure 6. Proposed model for nuage/piP‐body partitioning in BmN4 cells.
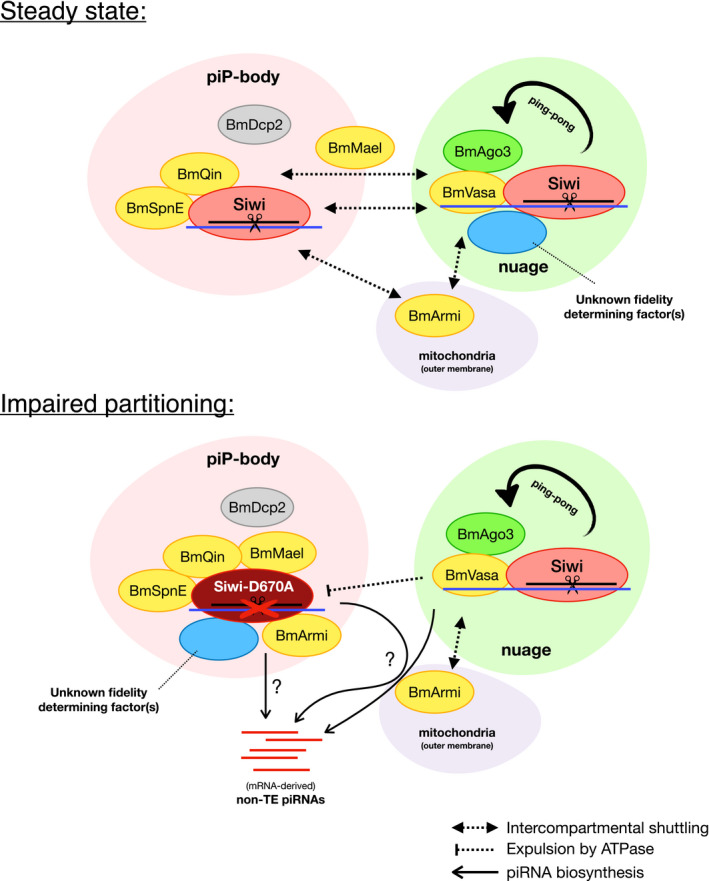
At steady state, nuage and piP‐body accommodate piRNA factors both stably localized at the condensate or transiently docked to it. Siwi, while does not accumulate in piP‐body, transiently enters piP‐body and interacts with BmSpnE and BmQin. An active slicer is required for Siwi to segregate from piP‐body and go back to nuage, where BmVasa uses its ATPase to remodel RNP interaction and modulates the ping‐pong cycle. Some Siwi‐interacting factors like BmMael and BmArmi can enter multiple compartments where Siwi is present. When Siwi‐D670A is expressed, the slicer mutant is expelled from nuage by RNP remodeling activity of BmVasa and is trapped in piP‐body while co‐aggregating with piP‐body factors, as well as those freely shuttling between compartments. A hypothetic fidelity determining factor could be similarly trapped in piP‐body by the mutant, resulting in the upregulation of non‐TE piRNAs. This production could be done in nuage, where native piRNA biosynthesis machineries may still function. Of note, the length distribution and the presence of 2′‐O‐methyl modification in non‐TE piRNAs suggest that these piRNAs are likely to be processed on the outer mitochondrial membrane and in the cytoplasm. It is also possible that non‐TE piRNA is directly generated from piP‐body, where a battery of piRNA biosynthesis factors abnormally accumulated.
