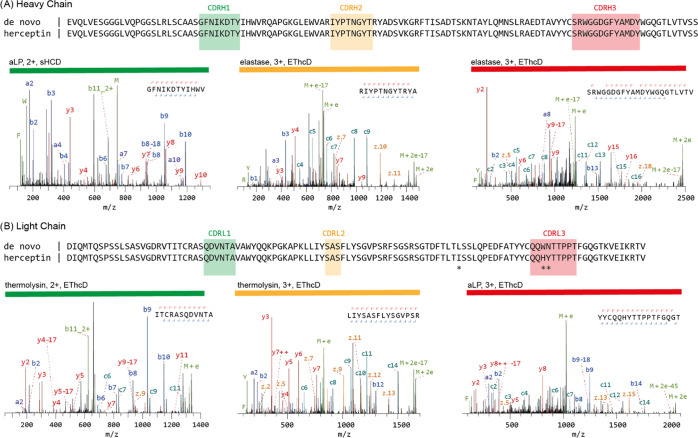
Figure 1.
Mass spectrometry-based de novo sequencing of the monoclonal antibody herceptin. The variable regions of the heavy (A) and light chains (B) are shown. The MS-based sequence is shown alongside the known herceptin sequence, with differences highlighted by asterisks (*). Exemplary MS/MS spectra supporting the assigned sequences of the heavy- and light-chain CDRs are shown below the alignments with protease, precursor charge state, and fragmentation type indicated. Peptide sequence and fragment coverage are indicated on top of the spectra, with b/c ions indicated in blue/teal and y/z ions in red/orange. The same coloring is used to annotate peaks in the spectra, with additional peaks such as intact/charge reduced precursors, neutral losses, and immonium ions indicated in green. Note that to prevent overlapping peak labels, only a subset of successfully matched peaks are annotated.