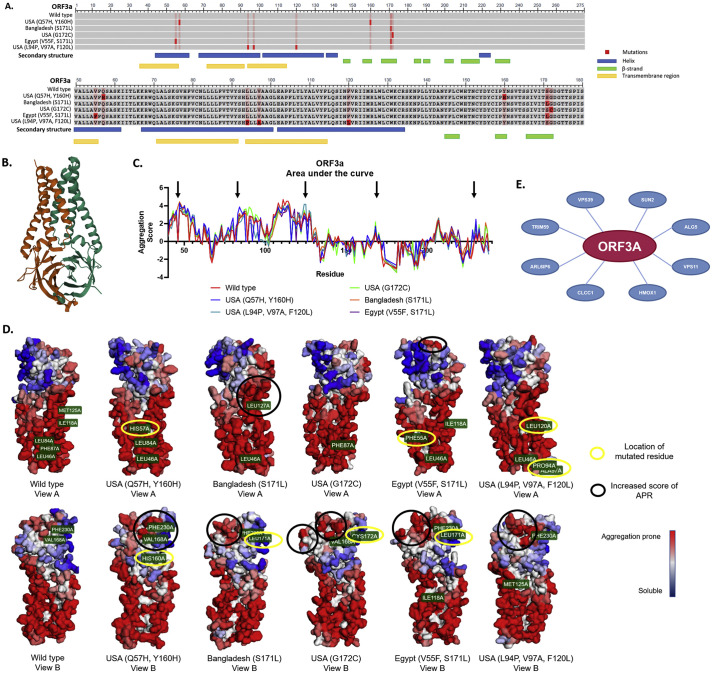
Fig. 8.
ORF3a variant structural analysis. a) COBALT sequence alignment analysis of the mutant variants vs. wild type. The bright red boxes represent the mutation in each variant and the light red boxes represent the residue that was changed in another variant. Below the sequence, the important characteristics of the secondary structure are shown; the blue boxes are alpha-helixes, the green boxes represent the beta-strands and the yellow boxes the transmembrane regions. b) Secondary structure of the ORF3a protein wild type reported in the PDB (6XDC). c) Area under the curve analysis of the aggregation score of each amino acid from each ORF3a variant. Black arrows point towards the peaks that showed a higher increase in the aggregation score compared to the wild type. d) Aggrescan 3D 2.0 reconstruction of the variant structures. Yellow circles show where the mutated amino acid is located, black circles show the important changes in the aggregation prone regions. e) CoVex Protein-Protein interaction network analysis. Red circle is the viral protein, blue circles are the host proteins. The interaction prediction tool is based on the human cell line HEK-293 T.