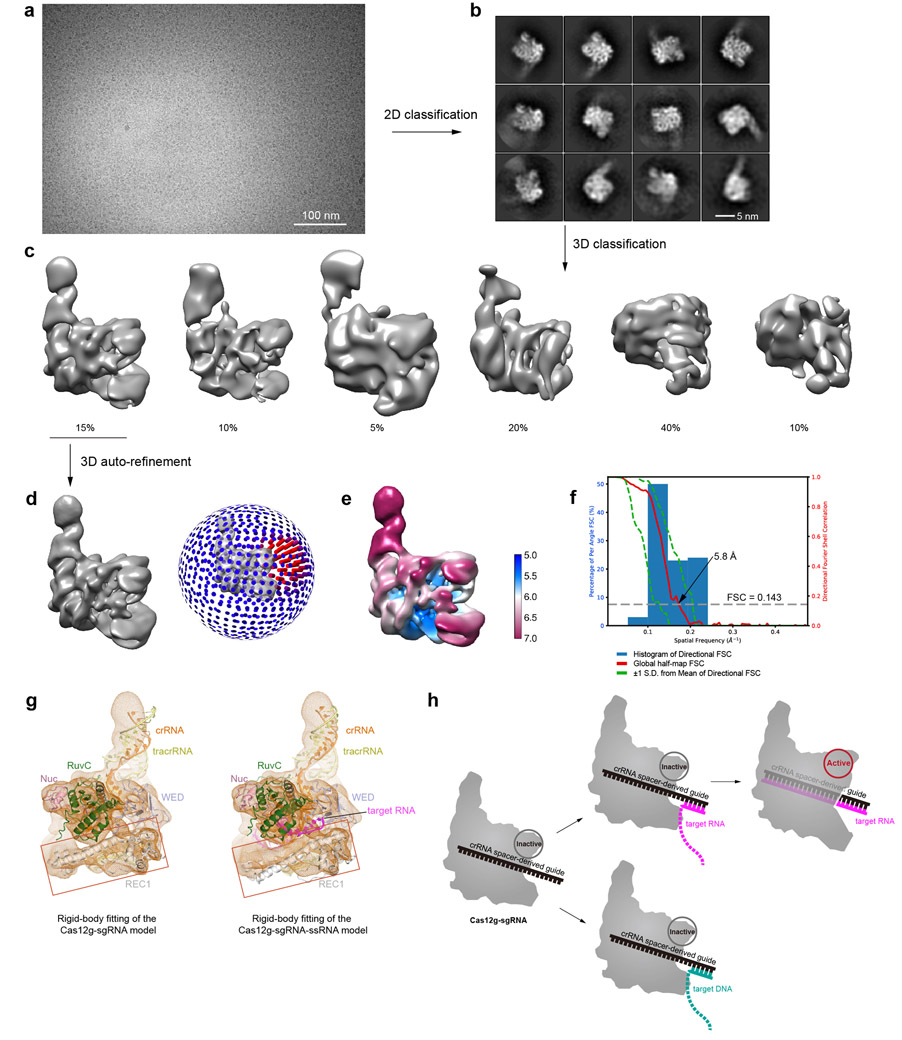
Extended Data Fig. 9. Cryo-EM analysis for the Cas12g-sgRNA-target DNA complex.
a, A representative raw cryo-EM micrograph of the Cas12g-sgRNA-target DNA complex from a total of 1083 micrographs. b, Representative 2D class averages from a total of 100 images. c, 3D classification. Classes 2-6 did not result in good reconstructions and likely represent damaged particles. d, 3D refinement for Class 1 with angular distribution. e, Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC). FSC plot for the reconstruction suggests an average resolution of 5.8 Å. f, Local resolution map. g, Comparison of the rigid-body fitting between the Cas12g-sgRNA complex and the Cas12g-sgRNA-target RNA complex. Red squares indicate the fitting of the REC1 domain, which shows that the model of the Cas12g-sgRNA complex fits better to the map (left panel). h, Schematic model for Cas12g activation. Target DNA fails to form fully assembled duplex required for Cas12g activation.