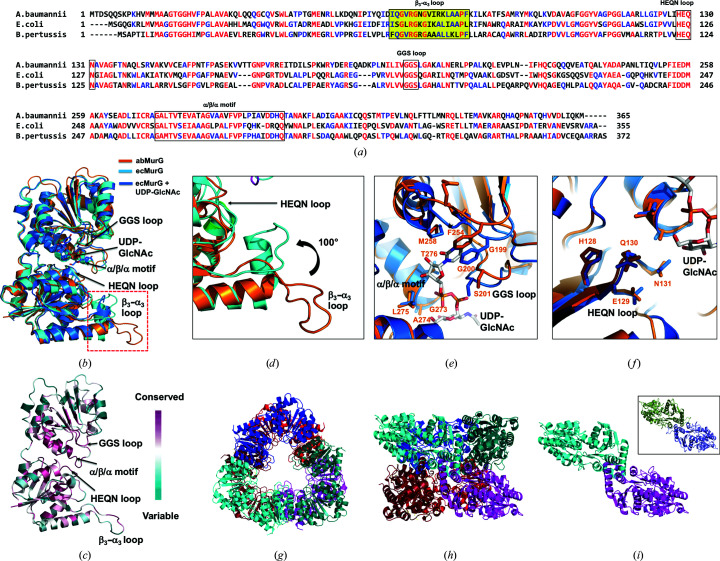
Figure 4.
Structure and sequence comparison of abMurG and ecMurG. (a) Sequence alignment of MurGs from different species. Mostly conserved and partially conserved residues are shown in red and blue, respectively. The HEQN loop, GGS loop and α/β/α motif are indicated with black boxes. The position of the β3–α3 loop is highlighted. (b) Structural superposition of abMurG (orange) with ecMurG (cyan) and the ecMurG–UDP–GlcNAc complex (blue). The red dotted box indicates the β3–α3 loop region. (c) Cartoon representation of abMurG colored according to the degree of amino-acid sequence conservation. The HEQN loop, GGS loop and α/β/α motif regions, which are critical for the accommodation of substrates in the structure of abMurG, are indicated. (d) Different structural details of the β3–α3 loop region between abMurG (orange) and ecMurG (cyan). (e) Superimposition of the structure of abMurG with ecMurG focusing on the UDP–GlcNAc-binding site. (f) Superimposition of the structure of abMurG with ecMurG focusing on the HEQN loop. (g, h) Putative hexameric structure of ecMurG modeled using the hexameric abMurG structure as a template: top (g) and side (h) views. (i) Type 1 interaction between two molecules in the hexameric structure of ecMurG. The type 1 interaction in the hexameric structure of abMurG is shown in a black box for comparison.