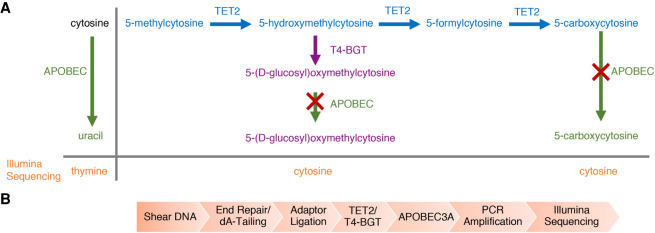
Figure 2.
Enzymatic methyl-seq (EM-seq) mechanism of action and workflow. (A) Principle pathways that are important for enzymatic identification of 5mC and 5hmC using EM-seq. The actions of TET2 (blue) and T4-BGT (purple) on 5mC and its oxidation products, as well as the activity of APOBEC3A (green) on cytosine, 5gmC, and 5caC are shown. The red cross represents no APOBEC3A activity. T4-BGT glucosylates 5hmC (pre-existing 5hmC and that formed by the action of TET2). TET2 converts 5mC through the intermediates 5hmC and 5fC into 5caC. APOBEC3A has limited activity on 5fC and undetectable activity on 5gmC and 5caC (Supplemental Fig. 3C). Uracil is replaced by thymine during PCR and is read as thymine during Illumina sequencing. (B) DNA is sheared to ∼300 bp, end repaired, and 3′-A-tailed. EM-seq adaptors are then ligated to the DNA. The DNA is treated with TET2 and T4-BGT before moving to the deamination reaction. The library is PCR amplified using EM-seq adaptor primers and can be sequenced on any Illumina sequencer.