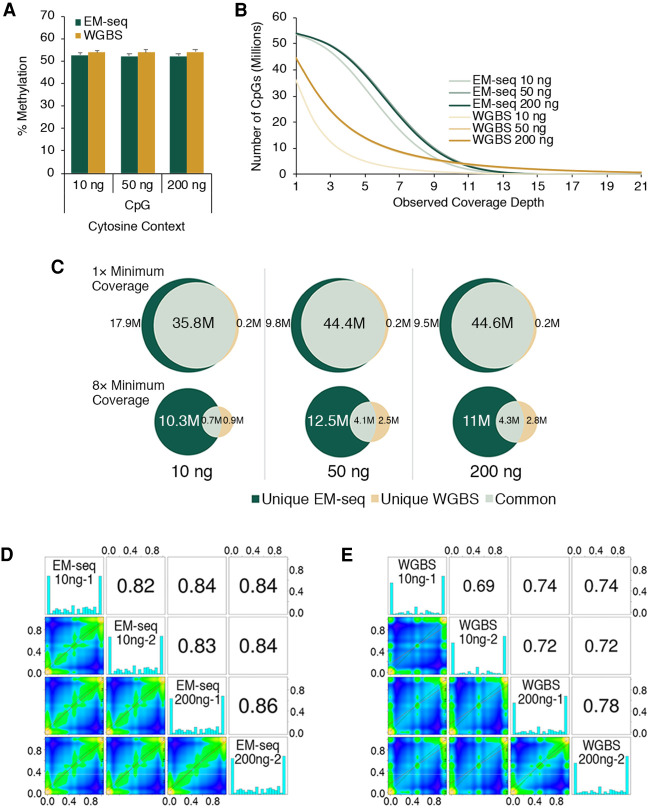
Figure 4.
EM-seq accurately represents methylation. Illumina NovaSeq data for 10, 50, and 200 ng of NA12878 DNA EM-seq and WGBS libraries were generated; 324 million paired reads for each library were aligned to a human + control reference genome using bwa-meth 0.2.2, and methylation information was extracted from the alignments using MethylDackel (https://github.com/dpryan79/MethylDackel). The top- and bottom-strand CpGs were counted independently, yielding a maximum of 56 million possible CpG sites. (A) NA12878 EM-seq and WGBS methylation in CpG contexts are similarly represented. The methylation state for NA12878 DNA in the CHG and CHH contexts and the unmethylated lambda control and CpG methylated pUC19 control DNAs are shown in Supplemental Figure 7. (B) The number of CpGs covered for EM-seq and bisulfite libraries was calculated and graphed at minimum coverage depths of 1× through 21×. (C) The number of CpGs detected was compared between the EM-seq and bisulfite libraries at 1× and 8× coverage depths. CpGs unique to EM-seq libraries or bisulfite libraries or those that were common to both are represented in the Venn diagrams. methylKit analysis at minimum 1× coverage shows good CpG methylation correlation between libraries made using 10 ng and 200 ng of NA12878 DNA for EM-seq (D) and WGBS (E) libraries. Methylation level correlations between inputs and replicates of EM-seq libraries are better than for WGBS libraries. Correlation between EM-seq and WGBS libraries at 10-ng, 50-ng, and 200-ng NA12878 DNA inputs are shown in Supplemental Figure 9D.