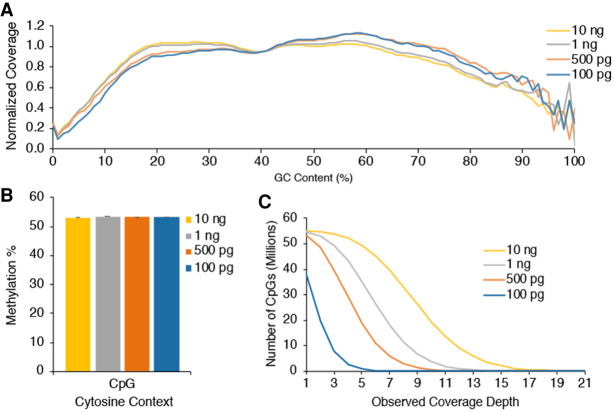
Figure 6.
Low-DNA-input EM-seq libraries. Low-input EM-seq libraries were made using 100 pg, 500 pg, 1 ng, and 10 ng of NA12878 DNA. Libraries were sequenced on a NovaSeq 6000 and evaluated for consistency before combining technical replicates and sampling 810 million total reads from each library type for methylation calling. (A) GC-bias plot for EM-seq library replicates shows even GC distribution. (B) CpG methylation levels are as expected for all DNA inputs at ∼53%. (C) CpG coverage as a function of minimum coverage depth is similar to standard-protocol libraries and shows the impact of library complexity and duplication on coverage.