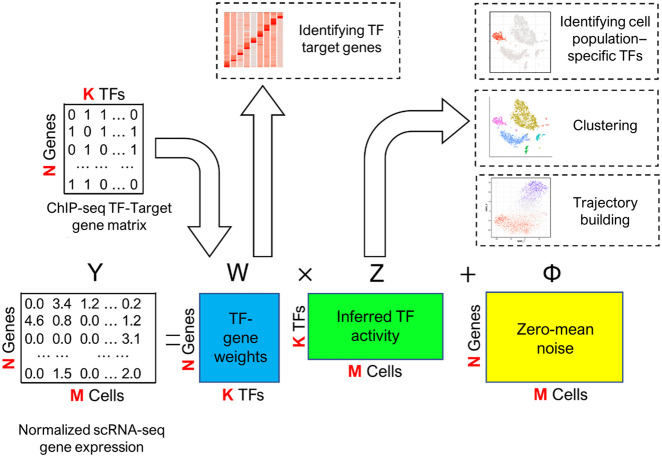
Figure 1.
Overview of the BITFAM model. The input to BITFAM is the log-normalized scRNA-seq data and a binary matrix with the predicted target genes for each transcription factor obtained from ChIP-seq data. A Bayesian factor analysis model with regulatory prior knowledge is built to learn matrices of transcription factor activities and transcription factor targets. The log-normalized scRNA-seq data, matrix Y, are decomposed to matrix W and matrix Z. The ChIP-seq data are incorporated as different prior distributions in matrix W. The posterior distributions of matrix W and matrix Z are inferred by the variational method (Ghahramani and Matthew 2000; Wainwright and Jordan 2008). The final matrices of W and Z used for downstream analysis are constructed by taking the means of 300 random samples from the posterior distributions. The matrix W is used to identify the target genes of transcription factors in the specific data set. The matrix Z is used in clustering and trajectory analysis as well as providing insights into the transcription factor regulatory activities.