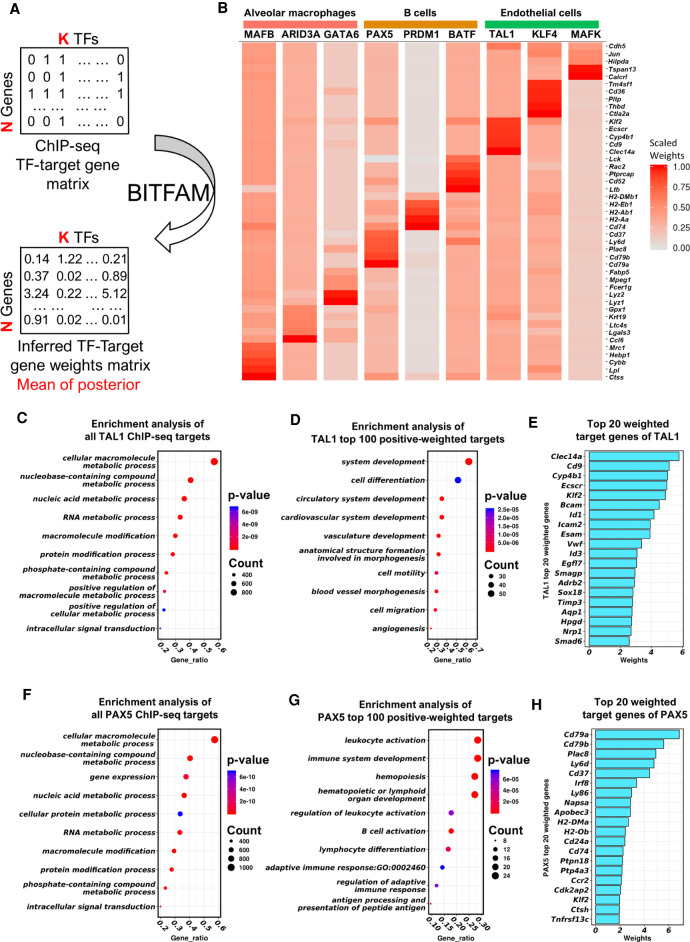
Figure 3.
BITFAM generates a ranking of preferred transcription factor target genes using scRNA-seq data. (A) The weights between the transcription factors and target genes inferred by BITFAM are based on ChIP-seq data and scRNA-seq data and identify preferred target genes in the specific data set. (B) Heatmap of five top-weighted target genes of transcription factors learned from Tabula Muris lung data. For each cell type, we depict TFs that were identified by BITFAM to be highly active in a given cell type. We list the top-weighted target genes based on the weights in the W matrix learned from the model and which are rescaled to [0,1] within each TF to generate the heatmap. (C) The top 10 significant GO terms of TAL1 ChIP-seq target genes in the Tabula Muris lung data. (D) The top 10 significant GO terms of TAL1 top 100 positive weighted genes learned from the Tabula Muris lung data. (E) The top 20 weighted target genes of TAL1. (F) The top 10 significant GO terms of PAX5 ChIP-seq target genes in the Tabula Muris lung data. (G) The top 10 significant GO terms of PAX5 top 100 positive weighted genes learned from the Tabula Muris lung data. (H) The top 20 weighted target genes of PAX5.