Abstract
Coronary artery disease (CAD) remains a major cause of mortality and morbidity worldwide. The aggregation of activated platelets on a rupturedatherosclerotic plaque is a critical step in most acute cardiovascular events like myocardial infarction. Platelet aggregation bothat baseline and after aspirin is highly heritable. Genome-wide association studies (GWAS) have identified a common variant within the first intron of the platelet endothelial aggregation receptor1 (PEAR1), to be robustly associated with platelet aggregation. In this study, we used targeted deep sequencing to fine-map the prior GWAS peak and identify additional rare variants of PEAR1that account for missing heritability in platelet aggregation within the GeneSTAR families.
1709 subjects (1043 EuropeanAmericans, EAand 666 African Americans, AA)from families in the GeneSTAR study were included in this study. In vitro platelet aggregation in response to collagen, ADP and epinephrine was measured at baseline and 14 days after aspirin therapy (81 mg/day). Targeted deep sequencing of PEAR1 in addition to 2kb of upstream and downstream of the gene was performed. Under an additive genetic model, the association of single variants of PEAR1with platelet aggregation phenotypeswere examined.Additionally, we examined the association between the burden of PEAR1 rarenon-synonymous variants and platelet aggregation phenotypes.
Of 532 variants identified through sequencing,the intron 1 variant, rs12041331, was significantly associated with all platelet aggregation phenotypes at baseline and after platelet inhibition with aspirin therapy. rs12566888, which is in linkage disequilibrium with rs12041331, was associated with platelet aggregation phenotypes but to a lesser extent. In the EA families,the burden of PEAR1missense variants was associated with platelet aggregation after aspirintherapy when the platelets were stimulated with epinephrine (p = 0.0009) and collagen (p = 0.03).In AAs, the burden of PEAR1 missense variants was associated, to a lesser degree, with platelet aggregation in response to epinephrine (p = 0.02) and ADP (p = 0.04).
Our study confirmed that the GWAS-identified variant, rs12041331, is the strongest variantassociated with platelet aggregation both at baseline and after aspirin therapy in our GeneSTAR families in both races. We identified additional association of rare missense variants in PEAR1 with platelet aggregation following aspirin therapy. However, we observed a racial difference in the contribution of these rare variants to the platelet aggregation, mostlikely due to higher residual missing heritability of platelet aggregation after accounting for rs12041331 in the EAs compared to AAs.
Keywords: platelet aggregation, PEAR1, deep targeted sequencing
Introduction:
Despite significant progress in prevention and treatment of cardiovascular diseases over the past several decades, atherosclerotic cardiovascular diseases (ASCVD) remainthe leading cause of mortality and morbidity worldwide1. The aggregation of activated platelets on a ruptured atherosclerotic plaque is one of the critical steps in thrombus formation during cardiovascular events like myocardial infarction, stroke and acute limb ischemia. Platelet aggregation is a tightly controlled process2and it is a delicate balance between endogenous platelet aggregation stimuli like ADP, collagen and epinephrine and endogenous platelet inhibitors like nitric oxide and prostacyclins3. Low dose aspirin (acetylsalicylic acid, ASA) has long been used for platelet inhibition in individuals with history of cardiovascular disease and now is considered to be the cornerstone of secondary prevention of ASCVD4. Platelet aggregation displays a high inter-individual variability at baseline and in response to anti-platelet drugs like aspirin. High baseline and on-treatment residual platelet activity has been linked to adverse cardiovascular outcomes including myocardial infarction5.The inter-individual variability of platelet reactivity does have a significant genetic component with the heritability estimates between 40% to60%6,7. The Genetic Study of Atherosclerosis Risk (GeneSTAR) is a family-based cohort, specifically designed to examine genetic elements of platelet aggregation in apparently healthy subjects with a family history of premature coronary artery disease.
Prior gene based8and genome wide9–11association studies have identified the platelet endothelial aggregation receptor-1 (PEAR1) locus to be associated with agonist induced platelet aggregation at baseline and after treatment with ASA. Our group previously reported that a common variant (rs12041331) in intron 1 of PEAR1was the peak GWAS-identified association signal between the PEAR1 locus and platelet aggregation phenotypes in the GeneSTAR families, in both African Americans (AA)11and European Americans (EA) 5,12. Additionally, variants in PEAR1 have been associated with platelet response to other pharmacological platelet inhibitors like clopidogrel13, prasugrel14and potentiallyclinical cardiovascular outcomes5,13. Furthermore, previous limited direct sequencing and imputation of PEAR1 coding regionsin GeneSTAR participants showedthere wasan association between burden ofPEAR1 synonymous variants and platelet aggregation15. PEAR1 is a novel platelet transmembrane proteininvolved in platelet contact-induced activation. Upon activation, PEAR1 becomes phosphorylated in a Src kinase-dependent manner leading to downstreamPI3K/Akt pathway activation. This cascade ultimately results in stabilization of platelet aggregates through glycoprotein αIIbβ.16,17
Despite these successes in understanding the pathophysiology and genetic bases of hemostasis, much of the heritability of platelet aggregation remains unexplained18. In this study, we sought to 1) refine/fine-mapthe prior GWAS peak inthe PEAR1 locus, i.e. attempt through sequencing to identify the causal variant for the GWAS-identified PEAR1 locus;2) identify common and rare variants in the PEAR1 locus;and 3) examine the individual (single-variant) and collective (burden of rare variants) association of the identified variants witha wide range of platelet aggregation phenotypes. Here, we extend our prior sequencing efforts in 104 subjects to allcoding and non-codingregions of PEAR1in a larger sample of 1,709 subjects from GeneSTAR families.
Methods:
Study subjects.
The study was approved by the Johns Hopkins Medicine Institutional Review Board. All participants provided written informed consent. GeneSTAR is a longitudinal prospective study of family members of index cases who were hospitalized with anacute coronary syndrome at age less than 60 years of age. The hospitalized index cases were used to access apparently healthy siblings and offspring whose had a family history of early onset coronary artery disease; in addition, the co-parents of the offspring were recruited. The study described here includes 1709 participants who participated in a two-week aspirin study and who had sequencing data.
All participants were between the ages of 21 and 80. Exclusion criteria included: history of allergy to aspirin, Hematocrit less than 30%, platelet count < 100,000/uL, white blood count > 20,000.uL, any bleeding disorder or any hemorrhagic event in the past (stroke, gastrointestinal bleed), use of any anticoagulants or anti-platelet agents (i.e. warfarin, persantin, clopidogrel), chronic or acute nonsteroidal anti-inflammatory agents (i.e COX-2 inhibitors that could not be discontinued), recent active gastrointestinal disorder, current pharmacotherapy for a gastrointestinal disorder, pregnancy or risk of pregnancy during the treatment trial, recent menorrhagia, known aspirin intolerance or allergic side effects, serious medical disorders (like autoimmune diseases, renal or hepatic failure, cancer or HIV-AIDS), chronic or acute use of glucocorticosteroid therapy or any drug that may interfere with the measured outcomes, serious psychiatric disorders, and, unable to independently make a decision to participate.Participants were instructed to avoid non-steroidal anti-inflammatory drugs 10 days prior to baseline measurements. Participants were then instructed to take 81 mg aspirin daily for 14 days.
Platelet aggregation.
Platelet function was assessed at baseline and after 2 weeks of aspirin therapy (81 mg/day). Using differential centrifugation of 3.2% citrated blood samples, platelet rich plasma (200,000 platelets/μl) was prepared. Optical aggregation was measured in a PAP-4 Aggregometer (Bio-Data Corp., Horsham, PA) after stimulating samples with collagen (2 μg/ml), adenosine diphosphate (10 μM), or epinephrine (2 μM) as described before15. Peak aggregation within 5 minutes of agonist stimulation was recorded as percentage between 0 and 100%. Amongst available platelet related phenotypes in GeneSTAR families, these phenotypes were chosen since prior GWAS and functional studies9,12,19were performed specifically under these conditions.
DNA sequencing and quality control.
DNA was extracted using standard protocols. Using Illumina HiSeq2000, targeted deep sequencing (>40x) of PEAR1 was performed at the Center for Inherited Disease Research (CIDR) of Johns Hopkins University. Target capture was designed by standard Agilent protocol for a total of 25kb of sequence representing the entire PEAR1 gene (Chr1:156,863,523–156,886,226 NCBI build 37, hg 19) in addition to 2 kb of up and downstream. DNA fragmentation was performed on 100ng-1ug of genomic DNA using a Covaris E210 system and libraries were then prepared.The Agilent Bionanalyzer (HiSensitivity) was used for quality control (fragment size and DNA quality). The high quality next-generation sequencing (NGS) data were processed (e.g., quality control (QC) and variant calling) via an automated pipeline CIDRSeqSuite 2.0, a set of software tools designed to perform secondary and initial tertiary analysis on NGS data from the Illumina HiSeq instrument. Variants were annotated usingANNOVAR software20for 1) position based on hg19 genome 2) functional relevance including intronic, synonymous, non-synonymous, insertion/deletion(indel), splice site, 5’ UTR, 3’UTR, downstream and upstream3) novelty based on presence in dsSNP147, and 4) rarity which is defined by minor allele frequency less than 5% in our sequencing data. The functional consequence/ deleteriousness of variants were predicted in-silico using SIFT21, Polyphen22, FATHMM23 and CADD24 models.
Statistical analysis.
All variants passing quality control (depth of coverage >7 and genotype quality > 20) were included in the data analysis (n=532 variants). Additional QC on each variant was performed including an assessment of deviation from the Hardy-Weinberg equilibrium. Only bi-allelic variants were retained for downstream analysis. 1709 participants were included for downstream association analyses including 12 subjects with coronary artery disease.Single variant association tests were done separately within each race using single variant score testing via RAREMETALWORKERsoftware. Meta-analysis of single variants across races was then performed using RAREMETAL software25. In the association models, we used age and sex adjusted residuals with family structureand top 2 principal components (PCs)from EIGENSTRAT as covariates as described before15. Correlation between variants in the PEAR1 locus is not negligible as noted in the linkage disequilibrium presented in Supplementary Figure 1. To address the issue of multiple testing accounting for the non-independence between the SNPs generated through the sequencing, we used LD-based type I error correction. We determined the number of independent SNPs at a pairwise LD of r2<0.8. Single-variant p-values of 0.00028 (0.05/175) and 0.00021(0.05/236) were considered significant in EAs and AAs,respectively. In the meta-analysis, we applied a Bonferroni correction and a p-value of 9.3×10−5 (0.05/532) was considered to be significant.
In addition to single variant analysis, we applied the SKAT gene-based collapsing method to increase power to identify the association of rare exonic mutational loadwith platelet aggregation phenotypes. Two sets of exonic variants were constructedand their cumulative effect was tested in multivariate analysis in which age, gender, family structure and PCs 1–2were included as covariates.The first set (Set1) was composed of all non-synonymous, stop gain, stop loss exonic variants (Set 1: Number of variants: 16 in EAs and 26 in AAs). Asecond set (Set2) was constructed from Set 1 by filtering out the variants that were not novel (present in dbSNP) or were not rare (MAF ≥ 0.05) in the Thousand Genomes Project database(Set 2: Number of variants: 14 in EAs and 23 in AAs). Therefore, Set 2 is a subset of novel and rare variants curated from Set 1. Finally,the SKAT test was repeated leaving out one variant at the time. This series of leave-one-outtests was done to identify the individual variant(s) that best explain the PEAR1SKAT p-value. If one variant had more contribution to the SKAT p-value, we would expect to see a more dramatic drop in p-value when the variant was left out.
All statistical analyses and models were performed using Scipyand Matplotlibpackages of python and RAREMETAL software25. Single SNP tests conditioning on peak SNPs within the region were all performed within RAREMETAL.
Results:
Patient characteristics.
The final cohort was composed of 1043 European Americans (EAs) and 666 African Americans (AAs). The mean age of EAswas 45 (SD 12.91), 457 (43.8%) were male and 8 (0.76%) had a history of cardiovascular event. The mean age of AAs was 43 (SD 12.33), 256 (38.4%) were males and 4 (0.6%) had a history of cardiovascular event (Table 1).
Table-1.
Demographics of subjects in each cohort.
| Characteristics | European Americans | African Americans |
|---|---|---|
| No. | 1043 | 666 |
| Age, years, (mean+/−SD) | 45.12 +/− 12.91 | 43.78 +/− 12.33 |
| Malesex, no. (%) | 457(43.8) | 256(38.4) |
| History of CAD, no. (%) | 8(0.7) | 4(0.6) |
Sequencing discoveries.
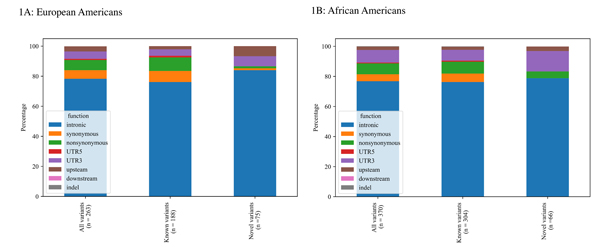
A 25kb region including PEAR1+/− 2kb upstream was sequenced and 532 biallelic single nucleotide variants that passed all quality control filters were identified. In EAs, 263 variants were identified (Figure 1A) of which60(22%) were common and 203 (78%) were rare. Among the 263 variants discovered in the EAs, 75 (28.5%) variants had not been reported in the public repositories (novel based on dbSNP147). In the AAs, 370 variants were identified(Figure 1B). Similar to the EAs, there were more rare than common variants (284vs 86, respectively) and 66 (18%)were considered to be novel. Thecomplete list of novel variants can be found in the supplementary materials (excel file).
Figure 1A-B:
Landscape of identified single nucleotide variants in the PEAR1 locus in European Americans (1A) and African Americans (1B) of GeneSTAR families.
Single variant association tests confirmedrs12041331as the likely causal variant of the GWAS signal.
Single variant tests were performed betweenall PEAR1variants and the range of platelet aggregation phenotypes separately in the EAs and AAs.Supplementary Figure 2 shows the results of PEAR1single variant association tests and phenotypes in each ethnic group.
In the AAs from GeneSTAR (Supplementary Table 1), thepreviously reported 11,12common intronicvariant (rs12041331, MAF = 0.391) was found to be significantly associated with platelet aggregation in response to collagen, ADP and epinephrine at baseline (p = 8.0×10−7, 1.02×10−7, 3.2×10−12, respectively). This association persisted after platelet inhibition with ASA (p = 2.0×10−5, 3.6×10−10, 6.1×10−15, respectively). rs12566888, which is in moderate linkage disequilibrium with rs12041331 (pairwise r2 = 0.46 in GeneSTAR AAs), reached statistical significance forplatelet aggregation in response to epinephrine at baselineand after aspirin therapy (p = 4.4×10−7, 6.8×10−8 respectively). However, this SNP was lower in significance compared to rs12041331. In theEAs (Supplementary Table 1), rs12041331 (MAF= 0.096) was associated with platelet aggregation in response to epinephrine at baseline (p = 3.9×10−5) and platelet aggregation in response to collagen and epinephrine (p = 7.6×10−6, 6.6×10−8, respectively) after ASA. No other variant crossed the significance thresholds for the single variants tests in the EAs. Also shown in Supplementary Table 1 is the locus specific heritability for these variants;the heritability of rs12041331 is consistently higher than the heritability due to rs12566888, and the heritability for rs12041331 is much higher in African Americans (range from 3.4% - 8.7%) compared to European Americans(range from 0.3 – 2.7%).
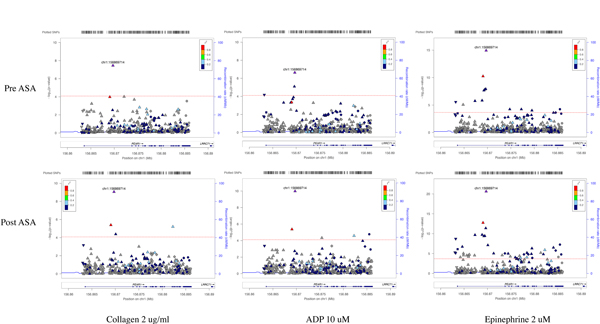
Figure 2 shows the results of meta-analysis of single variant association tests of platelet aggregation phenotypes, and the PEAR1 variantrs12041331 (chr1:156869714)remains the strongest single variant that determines platelet aggregation at baseline and after ASA. rs12566888 crossed significance thresholds forplatelet aggregation in response to epinephrine, collagen and ADP after ASA. This SNP was only associated with platelet aggregation in response to epinephrine at baseline. To hone in on the causal variant that explains this common variant association, and to test whether the association of rs12566888 with platelet aggregation phenotypes in this meta-analysis was truly independent of rs12041331, we performed conditional analysis. Conditioning on rs12041331, rs12566888 lost its association with platelet related phenotypes. However, conditioning on rs12566888, rs12041331 retained its association with platelet aggregation phenotypes (Supplementary Table 2). Additionally, the evidence for association with rs12041331 across all the platelet aggregation phenotypes is consistently stronger in the post ASA platelet aggregation for all agonists compared to baseline aggregation.
Figure 2:
Meta-analysis of association tests between PEAR1 variants with 3 platelet aggregation phenotypes in GeneSTAR families. Platelet aggregation was measured in response to collagen 2 ug/ml, ADP 10 uM and epinephrine 2 uM at baseline (pre-ASA) and 14 days after aspirin therapy (post-ASA). rs12041331 (purple triangle chr1:156869714) was significantly associated with all platelet aggregation related phenotypes at baseline and after aspirin therapy. Red lines reflect the Bonferroni threshold.
SKAT shows a significant association between exonic non-synonymous mutational burden of PEAR1 and platelet related phenotypes.
For the set-based SKAT analysis, two different sets of variants were constructed. The first set was composed of all non-synonymous, stop gain and stop loss exonic variants identified in the PEAR1 locus. (Set 1: Number ofvariants: 16 in EAs and 26 in AAs). We constructed a second subset from Set1 by filtering out the variants that were not novel (they were present in dbSNP147) or had MAF ≥ 0.05 in the Thousand Genomes Project database (Set 2: Number ofvariants:14 in EAs and 23 in AAs).
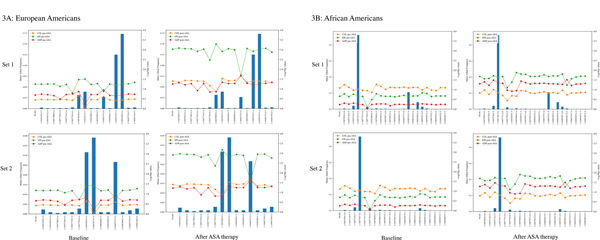
Table 2 shows the results of the SKAT test for association in each ethnic group. In EAs, we observeda significant SKAT signal of association with platelet aggregation in response to epinephrine and collagen after ASA(p = 0.0009, 0.039, respectively). We did not observe an association between mutational burden of PEAR1and platelet aggregation phenotypes at baseline. We then performed a leave-one-out analysis for SKAT to identify the individual variants that best explain the PEAR1 mutational burden (Figure3A–B). This analysisidentified four variants that contributed the most to the burden signal in EAs (Figure 3A). These 4 variants are Chr1:156877520 (rs149254521, MAF = 0.0009), Chr1:156877785 (rs757375280, MAF = 0.002), Chr1:156878044 (rs147639000, MAF = 0.03) and Chr1:156882757 (rs41299597, MAF = 0.02). We noted no LD between rs12041331 and these exonic variants, and these rare variants represent an independent contribution to platelet aggregation above the intronic common variant signal. Furthermore, we added rs12041331 to the constructed sets to evaluate the impact of rs12041331 within the gene-based framework of SKAT analyses. rs12041331 improved the SKAT p-values in EAs(Supplementary Table 3). This effect was more prominent in platelet aggregation in response to all three agonists after ASA. All four variants are missense variants resulting in an amino acid change in PEAR1, but only one variant, Chr1:156877785, was predicted to be damaging by Polyphen.
Table-2.
p-values of SKAT analysis of rare missense, stop-gain and stop loss variants in PEAR1 locus. Set 1 includes all missense, stop-gain and stop loss variants on PEAR1 locus. Set 2 was constructed from Set1 by excluding the variants that were not novel (they were present in dbSNP147) or had MAF ≥ 0.05 in the Thousand Genomes Project database.
| Ethnic group-phenotype | Set 1 | Set 2 |
|---|---|---|
| European Americans | ||
| ADP pre-ASA | 0.203 | 0.22 |
| ADP post-ASA | 0.056 | 0.06 |
| EPI* pre-ASA | 0.056 | 0.067 |
| EPI post-ASA | 0.00094 | 0.00121 |
| COL** pre-ASA | 0.363 | 0.379 |
| COL post-ASA | 0.039 | 0.039 |
| African Americans | ||
| ADP pre-ASA | 0.587 | 0.57 |
| ADP post-ASA | 0.049 | 0.051 |
| EPI pre-ASA | 0.225 | 0.215 |
| EPI post-ASA | 0.021 | 0.019 |
| COL pre-ASA | 0.075 | 0.072 |
| COL post-ASA | 0.16939 | 0.176 |
| Meta-analysis | ||
| ADP pre-ASA | 0.165 | 0.164 |
| ADP post-ASA | 0.032 | 0.032 |
| EPI pre-ASA | 0.149 | 0.157 |
| EPI post-ASA | 0.00082 | 0.00084 |
| COL pre-ASA | 0.071 | 0.07 |
| COL post-ASA | 0.249 | 0.261 |
EPI : epinephrine,
COL : collagen
Figure 3A-B:
Leave-one-out analysis to identify variants contributing the most to the burden test association signal in European Americans (3A) and African Americans (3B). Bars represent the minor allele frequency of variants and line-dots show the SKAT burden –Log (p-value) in absence of that specific variants. In European Americans, Chr1:156877520 (rs149254521), Chr1:156877785 (rs757375280), Chr1:156878044 (rs147639000) and Chr1:156882757 (rs41299597) contributed the most to the burden signal. In African Americans, Chr1:156877782 (rs145275734) has the most contribution to the burden signal.
Similarly, in African Americans, aggregated exonic variants were significantly associated with platelet aggregation in response to epinephrine and ADPafter ASA (p = 0.021, 0.049) but not collagen. Using the leave-one-out strategy(Figure3B), we found Chr1:156877782 (rs145275734, MAF = 0.02) explains the major part of SKAT signal in African Americans. This rare variant substitutes Glycine with Cysteine in amino acid 281 of PEAR1 (Gly281Cys). This change in PEAR1 protein structure was predicted to be damaging by Polyphen software. No LD between rs12041331 and rs145275734 has been reported in the Thousand Genomes Project (r2 = 0.01) or in our data (r2 = 0.01). Similar to the pattern noted in the EAs, we did not observe an association between nonsynonymous mutational burden of PEAR1 and platelet aggregation phenotypes at baseline in the AAs (Table 2).Unlike the EAs, rs12041331 did not improve the SKAT p-values when it was added to the constructed sets in the AAs(Supplementary Table 3).
Meta-analysis of SKAT tests of both races showed that the burden of non-synonymous, stop-gain and stop-loss variants was associated with platelet aggregation in response to epinephrine and ADP but not collagen after ASA (Table 2). No association was detected for platelet aggregation at baseline.
Discussion:
Atherosclerotic cardiovascular disease is associated with high morbidity, mortality and poor prognosis.ASCVD usually results from the activation, aggregation, and adhesion of platelets to eroded or ruptured atherosclerotic plaque and aspirin has long been used for primary and secondary prevention of ASCVD. Variability in platelet aggregation at baseline and in response to aspirin is highly heritable6,7and is strongly associated with the risk of myocardial infarction and stroke26,27. Our group has previously identified a common genetic variant in intron 1 of the PEAR1 gene, rs12041331, that accounts for 10% of native platelet function11,12. This locus accounts for a higher fraction (15%) of the total phenotypic variance in platelet function after aspirin exposure and potentially plays a larger role in platelet aggregation pathways that are independent of cyclooxygenase (COX). The PEAR1 gene comprises 23 exons and 22 introns, and its encoded protein participates in extracellular protein–protein interactions. PEAR1 is highly expressed on the surface of platelets and endothelial cells28. It contains 15 extracellular growth factor-type domains and both intracellular and extracellular zones of the PEAR1 protein can bind to phosphotyrosine28. Previous studies have explored the functional importance of PEAR1 in platelet degranulation and aggregation16,17. Kauskot et al 16showed PEAR1 stimulation with polyclonal antibodies against extracellular PEAR1 domain triggerssrc family kinase (SFK)-dependent phosphorylation of PEAR1leading to activation of PI3K/Akt signaling pathway. Vandenbriele et al 17showed Dextran Sulfateinducesfibrinogen receptor (αIIbβ3) activation via PEAR1-dependent (PEAR11/c-Src/PI3K/Akt) pathway resulting in stabilization of platelet aggregates. In this study, we embarked on deep targeted sequencing of the PEAR1locus to fine-map the GWAS peak and to look for additional variants that may explain the missing heritability of platelet aggregation phenotypes.
Single variant association tests confirmed rs12041331 as the strongestindependently associated variant with platelet aggregation phenotypes at baseline and after aspirin therapy in the GeneSTAR families. In our prior work9, an imputed version of rs12566888 (imputation quality, r2 = 0.982 in EAs and 0.8934 in AAs) appeared to be associated, to a lesser degree, with the platelet aggregation phenotypes in our GeneSTAR families; this is a variant that was reported to be the peak variant in the Framingham Heart Study from the prior GWAS. Here, we confirm with sequencing that rs12566888 retains its significance in GeneSTAR, but remains second in significance to rs12041331. It is important to note thatrs12566888 is in modest LD with rs12041331. Conditional analyses suggested the association signal of rs12566888 is not independent of rs12041331; i.e. the significance of rs12566888 is completely lost when rs12041331 is conditioned upon. In contrast, rs12041331 retained its significance even after conditioning upon rs12566888 suggesting an independent association with platelet aggregation phenotypes in the GeneSTAR families. Additionally,rs12041331, unlike rs12566888, has been linked directly to methylation-dependent expression of PEAR129. rs12041331 removes a cytosine guanine dinucleotide (CpG) site in an enhancer region within intron 1 where the G allele has been shown to bind with higher affinity to several nuclear proteins than the A allele29. Allele specific methylation of the G allele also marks higher methylation at a CpG island located at the untranslated region of PEAR1 containing binding sites for CTCF. It has been shown thatthe lower methylation at this CTCF binding region that is associated with the A allele of rs12041331 increased CTCF binding, and leads to lower PEAR1 expression 29. Taken jointly, our comprehensive query of the PEAR1 locus relying on sequencing and this prior functional data suggests that rs12041331 is the most likely causal variant for the PEAR1 locus in our families.
Examining ethnic groups separately, rs12041331 showed a larger effect size on allplatelet aggregation phenotypes in the African Americans compared to the European Americans(Supplementary Table1). These observations can bepartly explained by higher minor allele frequency (0.391 vs 0.096) and larger effect size of rs12041331 in the African Americans compared to the European Americans.Together, this results in much lower locus specific heritability in the European Americans compared to the African Americans.Furthermore, similar to the prior studies30,19, we observed that the effect of rs12041331 on platelet aggregation is agonist dependent in European Americans.
Generally rare variants individually do not have enough power to reach statistical significance in single variant association studies. One alternative approach is using collapsing techniques like SKAT to assess the cumulative effect of rare variants on the phenotypes. Although none of the exonic variants individuallyreached statistical significance, the burden of non-synonymous, stop-loss, stop-gain variants of PEAR1 was associated with the post ASA platelet aggregation phenotypes in the GeneSTAR EA families. In the GeneSTAR AAs, the PEAR1 burden was marginally associated with post ASA platelet aggregation. As noted above, the contribution of rs12041331 to the phenotype variance in the AAsis higher than in the EAs, and there is more residual unexplained missing heritability in the EA group. This may partially explain why SKATtests were more robustly associated with the phenotypes in the EA families and is along the line of the observations from a limited sample sequencing approach previously reported15. When we added rs12031441 to the constructed sets, we noted that rs12031441 improved the p-value of association only in EAs but not in AAs. This is expected given that the weight of each variant in SKAT model is an inverse function of MAF of the variant and more weight is placed on rare variants compared to common variants31.Here, we have lower MAF of rs12041331 in EAs compared to AAs(0.09 vs 0.39).The leave-one-out approach implicates 4 non-synonymousvariants contributing the most to the SKAT signal in the EAs and the lack of linkage disequilibrium between these four variants and rs12041331 suggests that this signal is not mediated by rs12041331. These 4 variants change the amino acid composition of PEAR1. Although PolyPhen2 predicted three out of these four variants are not damaging to the protein structure, the full functional effect of these variants on PEAR1 expression and function is yet to be discovered.
Whilemultiple studies have convincingly shown the contribution of PEAR1 variants toagonist induced platelet aggregation phenotypes9,11–13,15, the role of PEAR1 variants in cardiovascular events isconflicting13,32,33.Unfortunately, the GeneSTAR study is underpowered to discover an association between variants of PEAR1and cardiovascular events.Ongoing meta-analysis of multiple large cohorts such as those within the Trans-Omics for Precision Medicine (TOPMed) Program with available cardiovascular events and genotype data will address this question in the near future.
In this study, we consistently observed a stronger association of PEAR1variants, both for the single SNPs and for the burden tests of rare coding variants,with platelet aggregation phenotypes after aspirin therapy. This is similar to the pattern observed from previously reported common variants analyses. Aspirin is a potent irreversible inhibitor of COX-134, the enzyme responsible for converting arachidonic acid to thromboxane A2 in platelets35. The generation and release of thromboxane from activated platelets generates a positive feedback loop that contributes to phenotypic variance in platelet function. In the presence of aspirin, which largely eliminates variability in platelet phenotype directly related to thromboxane A2, PEAR1 variants account for a larger proportion of the remaining variance in platelet aggregation12. This finding is consistent with an impact of PEAR1 on aggregation mediated by collagen, epinephrine, and ADP, each of which is known to activate platelets through pathways that are only partly related to thromboxane generation6. Because PEAR1 variants contribute more to platelet phenotypes in the presence than absence of aspirin, it is reasonable to speculate that the clinical impact, if any, of PEAR1 variants on platelet-mediated thrombosis would be more apparent in a population on aspirin therapy.
In conclusion, this study implicates rs12041331 as the causal variant behind the GWAS identified locus in the GeneSTAR African American and European American families for platelet aggregation phenotypes. Additionally, it supportsan additional role of exonic non-synonymous variants in platelet aggregation.
Supplementary Material
Acknowledgements:
The authors are grateful to the study participants.
Declaration of Interest:
GeneSTAR was supported by grants from the National Institutes of Health/National Heart, Lung, and Blood Institute (U01 HL72518 and HL087698 to LC Becker; HL112064 to RA Mathias) and by a grant from the National Institutes of Health/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center.
References:
- 1.Berenson GS, Srinivasan SR, Bao W, Newman WP, Tracy RE, Wattigney WA, Newman WP. Association between multiple cardiovascular risk factors and atherosclerosis in children and young adults. The Bogalusa Heart Study. The New England Journal of Medicine. 1998;338(23):1650–6. doi: 10.1056/NEJM199806043382302 [DOI] [PubMed] [Google Scholar]
- 2.Hennekens CH, Dyken ML, Fuster V. Aspirin as a therapeutic agent in cardiovascular disease: A statement for healthcare professionals from the American Heart Association. Circulation. 1997;96(8):2751–2753. [DOI] [PubMed] [Google Scholar]
- 3.Jin RC, Voetsch B, Loscalzo J. Endogenous mechanisms of inhibition of platelet function. Microcirculation. 2005;12(3):247–258. doi: 10.1080/10739680590925493 [DOI] [PubMed] [Google Scholar]
- 4.Fuster V, Dyken ML, Vokonas PS, Hennekens C. Aspirin as a therapeutic agent in cardiovascular disease. Special Writing Group. Circulation. 1993;87(2):659–675. doi: 10.1161/01.CIR.87.2.659 [DOI] [PubMed] [Google Scholar]
- 5.Qayyum R, Becker DM, Yanek LR, Faraday N, Vaidya D, Mathias R, Kral BG, Becker LC. Greater Collagen-Induced Platelet Aggregation Following Cyclooxygenase 1 Inhibition Predicts Incident Acute Coronary Syndromes. Clinical and Translational Science. 2015;8(1):17–22. doi: 10.1111/cts.12195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faraday N, Yanek LR, Mathias R, Herrera-Galeano JE, Vaidya D, Moy TF, Fallin MD, Wilson AF, Bray PF, Becker LC, et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115(19):2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584 [DOI] [PubMed] [Google Scholar]
- 7.Bray PF, Mathias RA, Faraday N, Yanek LR, Fallin MD, Herrera-Galeano JE, Wilson AF, Becker LC, Becker DM. Heritability of platelet function in families with premature coronary artery disease. Journal of thrombosis and haemostasis : JTH. 2007;5(8):1617–23. doi: 10.1111/j.1538-7836.2007.02618.x [DOI] [PubMed] [Google Scholar]
- 8.Herrera-Galeano JE, Becker DM, Wilson AF, Yanek LR, Bray P, Vaidya D, Faraday N, Becker LC. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(8):1484–1490. doi: 10.1161/ATVBAHA.108.168971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, Tofler G, Lin SJ, Kraja AT, Province MA, Yang Q, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nature Genetics. 2010;42(7):608–613. doi: 10.1038/ng.604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eicher JD, Chami N, Kacprowski T, Nomura A, Chen MH, Yanek LR, Tajuddin SM, Schick UM, Slater AJ, Pankratz N, et al. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. American Journal of Human Genetics. 2016;99(1):40–55. doi: 10.1016/j.ajhg.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qayyum R, Becker LC, Becker DM, Faraday N, Yanek LR, Leal SM, Shaw C, Mathias R, Suktitipat B, Bray PF. Genome-wide association study of platelet aggregation in African Americans. BMC genetics. 2015;16:58. doi: 10.1186/s12863-015-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faraday N, Yanek LR, Yang XP, Mathias R, Herrera-Galeano JE, Suktitipat B, Qayyum R, Johnson AD, Chen MH, Tofler GH, et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood. 2011;118(12):3367–3375. doi: 10.1182/blood-2010-11-320788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JP, Ryan K, O’Connell JR, Horenstein RB, Damcott CM, Gibson Q, Pollin TI, Mitchell BD, Beitelshees AL, Pakzy R, et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circulation: Cardiovascular Genetics. 2013;6(2):184–192. doi: 10.1161/CIRCGENETICS.111.964627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang Q, Cui Y, Zhao X, Zhao N. Identification of PEAR1 SNPs and their influences on the variation in prasugrel pharmacodynamics. Pharmacogenomics. 2013;14(10):1179–89. doi: 10.2217/pgs.13.108 [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Suktitipat B, Yanek LR, Faraday N, Wilson AF, Becker DM, Becker LC, Mathias RA. Targeted Deep Resequencing Identifies Coding Variants in the PEAR1 Gene That Play a Role in Platelet Aggregation. PLoS ONE. 2013;8(5). doi: 10.1371/journal.pone.0064179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauskot A, Di Michele M, Loyen S, Freson K, Verhamme P, Hoylaerts MF. A novel mechanism of sustained platelet αIIbβ3 3 activation via PEAR1. Blood. 2012;119(17):4056–4065. doi: 10.1182/blood-2011-11-392787 [DOI] [PubMed] [Google Scholar]
- 17.Vandenbriele C, Sun Y, Criel M, Cludts K, Van kerckhoven S, Izzi B, Vanassche T, Verhamme P, Hoylaerts MF. Dextran sulfate triggers platelet aggregation via direct activation of PEAR1. Platelets. 2016;27(4):365–372. doi: 10.3109/09537104.2015.1111321 [DOI] [PubMed] [Google Scholar]
- 18.Johnson AD. The genetics of common variation affecting platelet development, function and pharmaceutical targeting. Journal of Thrombosis and Haemostasis. 2011;9(1 S):246–257. doi: 10.1111/j.1538-7836.2011.04359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng LL, Zhao YQ, Zhou ZY, Jin J, Zhao M, Chen XM, Chen LY, Cai YF, Li JL, Huang M. Associations of MDR1, TBXA2R, PLA2G7, and PEAR1 genetic polymorphisms with the platelet activity in Chinese ischemic stroke patients receiving aspirin therapy. Acta Pharmacologica Sinica. 2016;37(11):1442–1448. doi: 10.1038/aps.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16). doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics. 2013;(SUPPL.76). doi: 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, Day INM, Gaunt TR. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Human Mutation. 2013;34(1):57–65. doi: 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46(3):310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng S, Liu D, Zhan X, Wing MK, Abecasis GR. RAREMETAL: Fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30(19):2828–2829. doi: 10.1093/bioinformatics/btu367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krasopoulos G, Brister SJ, Beattie WS, Buchanan MR. Aspirin “resistance” and risk of cardiovascular morbidity: systematic review and meta-analysis. BMJ. 2008;336(7637):195–198. doi: 10.1136/bmj.39430.529549.BE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snoep JD, Hovens MMC, Eikenboom JCJ, van der Bom JG, Huisman M V. Association of laboratory-defined aspirin resistance with a higher risk of recurrent cardiovascular events: a systematic review and meta-analysis. Archives of internal medicine. 2007;167(15):1593–9. doi: 10.1001/archinte.167.15.1593 [DOI] [PubMed] [Google Scholar]
- 28.Nanda N, Bao M, Lin H, Clauser K, Komuves L, Quertermous T, Conley PB, Phillips DR, Hart MJ. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. Journal of Biological Chemistry. 2005;280(26):24680–24689. doi: 10.1074/jbc.M413411200 [DOI] [PubMed] [Google Scholar]
- 29.Izzi B, Pistoni M, Cludts K, Akkor P, Lambrechts D, Verfaillie C, Verhamme P, Freson K, Hoylaerts MF. Allele-specific DNA methylation reinforces PEAR1 enhancer activity. Blood. 2016;128(7):1003–1012. doi: 10.1182/blood-2015-11-682153 [DOI] [PubMed] [Google Scholar]
- 30.Backman JD, Yerges-Armstrong LM, Horenstein RB, Newcomer S, Shaub S, Morrisey M, Donnelly P, Drolet M, Tanner K, Pavlovich MA, et al. Prospective Evaluation of Genetic Variation in Platelet Endothelial Aggregation Receptor 1 Reveals Aspirin-Dependent Effects on Platelet Aggregation Pathways . Clinical and Translational Science. 2017;10(2):102–109. doi: 10.1111/cts.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. American Journal of Human Genetics. 2013;92(6):841–853. doi: 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voora D, Horton J, Shah SH, Shaw LK, Newby LK. Polymorphisms associated with in vitro aspirin resistance are not associated with clinical outcomes in patients with coronary artery disease who report regular aspirin use. American Heart Journal. 2011;162(1). doi: 10.1016/j.ahj.2011.03.026 [DOI] [PubMed] [Google Scholar]
- 33.Yang W-Y, Petit T, Cauwenberghs N, Zhang Z- Y, Sheng C- S, Thijs L, Salvi E, Izzi B, Vandenbriele C, Wei F- F, et al. PEAR1 is not a major susceptibility gene for cardiovascular disease in a Flemish population. BMC Medical Genetics. 2017;18(1):45. doi: 10.1186/s12881-017-0411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vane JR, Botting RM. The mechanism of action of aspirin. In: Thrombosis Research. Vol. 110. 2003. p. 255–258. doi: 10.1016/S0049-3848(03)00379-7 [DOI] [PubMed] [Google Scholar]
- 35.Naimo PS, McGiffin D, Konstantinov IE. Aspirin resistance in the era of personalized medicine: Should we not take it personally? Journal of Thoracic and Cardiovascular Surgery. 2015;150(6):e99–e100. doi: 10.1016/j.jtcvs.2015.09.049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.