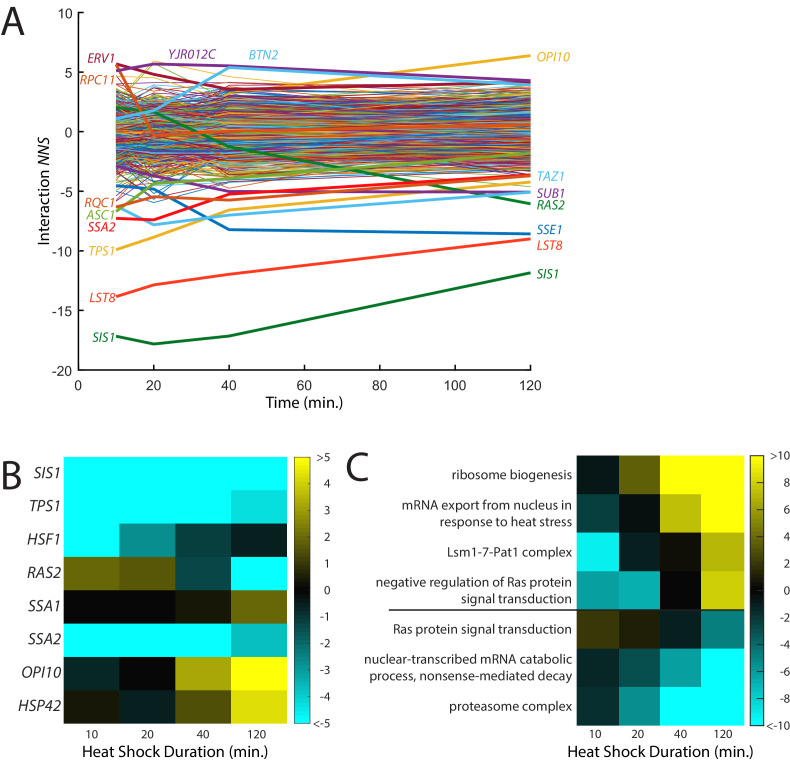
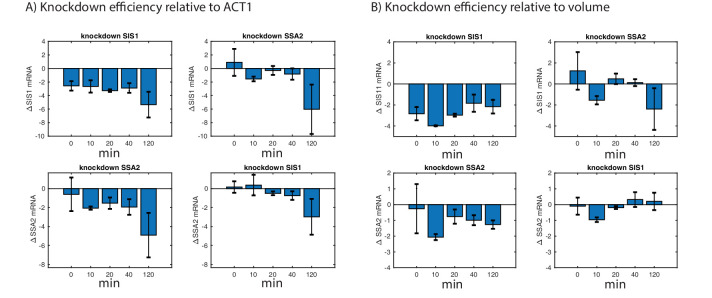
Figure 4. Time-resolved interactions between genetic knockdowns and heat stress.
(A) Time course of interaction NNS for each gene with 39°C heat shock. Time points were measured at 10, 20, 40, and 120 min. (B) Heat map of interaction NNS for selected genes in the screen. (C) Heat map of GO category scores for selected categories with differential effects in early and late time points.