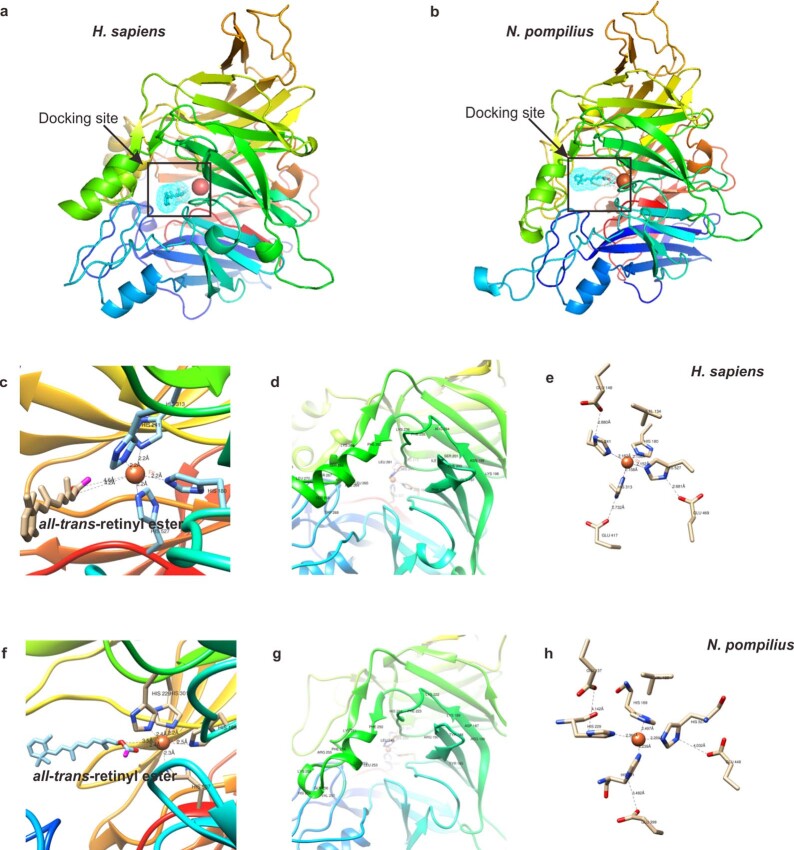
Extended Data Fig. 6. Modeling and docking of RPE65 and all-trans retinyl ester in N. pompilius and H. sapiens.
Structure model of H. sapiens RPE65 (a) and N. pompilius RPE65 (b) with all-trans retinyl ester, which located near the active site defined by the iron ion. The ion cofactor is found near the top face of the propeller axis and is conserved in H. sapiens and N. pompilius, which is directly coordinated by four His residues (His180, His241, His313, His527 in H. sapiens; His169, His229, His301, His507 in N. pompilius), with average bond length of 2.16 Å in H. sapiens, and 2.34 Å in N. pompilius. Ferrous iron is required for its catalytic activity, binding to the hydroxyl oxygen to catalyze the isomerization reaction. The docking site details were displayed, revealing that a shorter average bond length (2.95 Å) between atRE and ion cofactor in N. pompilius (Fig f), than that (4.4 Å) in H. sapiens (Fig c), suggesting the catalytic potential of N. pompilius RPE65. The hydropholic tunnel of N. pompilius RPE65, leads from the protein surface to active site, the mouth of which is surrounded by three groups of residues (185–190, 222–224, and 249–259, Fig g), highly conserved with that in H. sapiens RPE65 (196–202, 234–236, and 261–271, Fig d). On the other hand, the N. pompilius RPE65 also shows a distinguishable character: the iron cofactor, ordinated by four His residues, three second shell Glu residues and a Val residue, displays a more loose structure (Fig h) than that in H. sapiens RPE65 (Fig e), which shows no obvious interference to its catalytic activity.