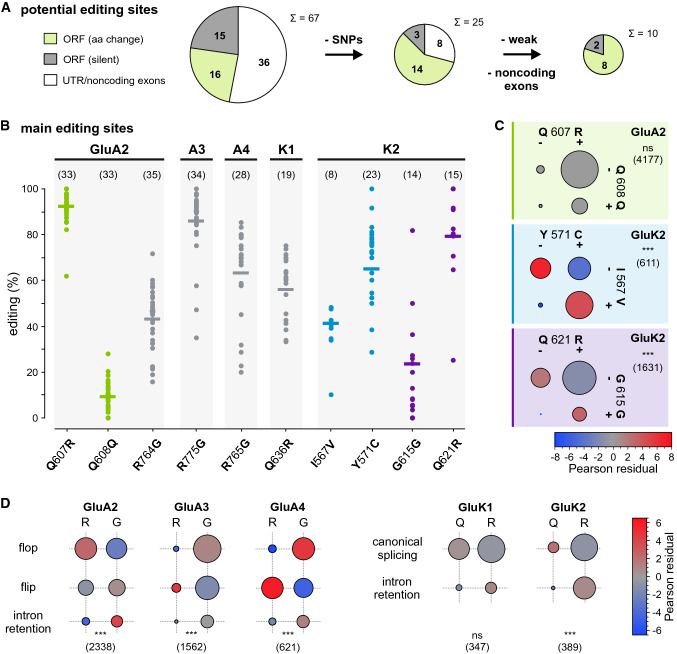
Fig. 5.
RNA editing in iGluRs and correlation with splicing. A Detection of single-nucleotide mismatches that report on potential A-to-I or C-to-U editing events (for details see Methods and Fig. S24). B Abundance of the ten most frequent events in different datasets (mean indicated by bars; (n) number of shown datasets (≥ 40 reads); see also Fig. S26). C Co-editing of sites within single-read distance. The circle sizes represent the read numbers obtained with editing-specific queries (see Table S8). Statistical testing shows independence for GluA2 Q607R/Q608Q editing, but interdependence for GluK2 Y571C/I567V and Q621R/G615G editing. D Relation between RNA editing and adjacent splice site usage. Statistical testing was performed with Pearson’s chi-squared test of independence (***p ≤ 0.0005; (n) number of total reads)