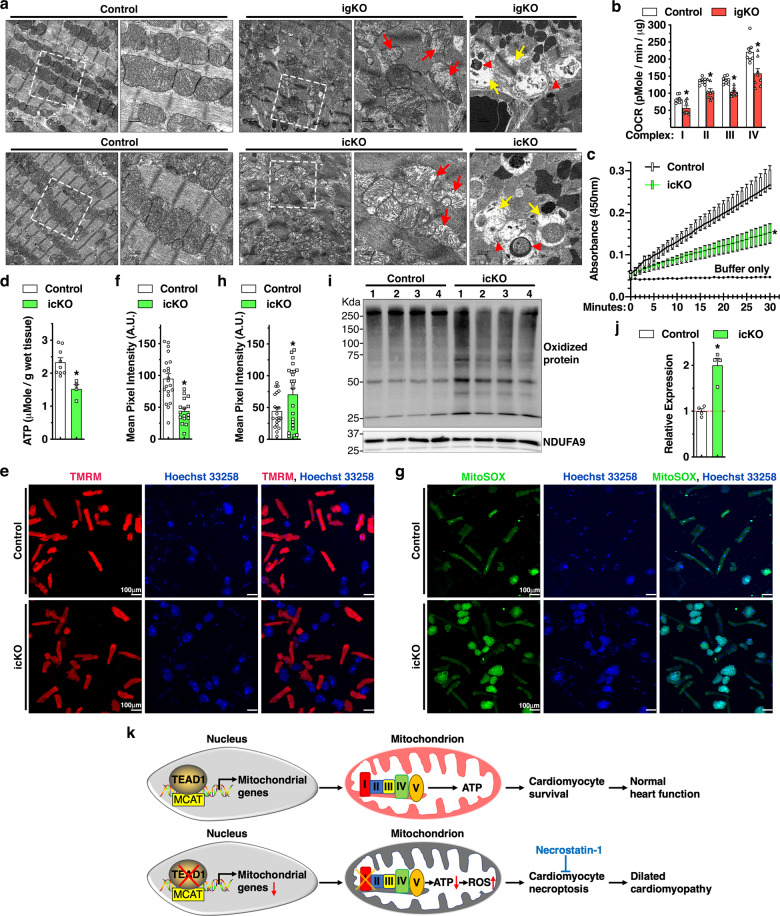
Fig. 7. Loss of Tead1 in CMs leads to mitochondrial dysfunction.
a Representative transmission electron microscopy images demonstrate mitochondrial morphology in control, Tead1 igKO (upper panel) and icKO (bottom panel) hearts. Red and yellow arrows indicate mitochondria that lost cristae and focal areas with myofibrillar lysis, respectively. Red arrow heads mark reminiscent of mitochondrial membrane or damaged mitochondria induced mitophagic vesicles. The boxed area is magnified on the right. b Mitochondrial bioenergetics assays were performed to measure oxygen consumption rate (OCR) in mitochondria isolated from control and Tead1 igKO hearts to determine ETC complex I-IV respiratory activity. N = 4. *p < 0.05. c Protein lysates were isolated from control and Tead1 icKO hearts. Equal amounts of protein were then used to determine complex I activity following the oxidation of NADH to NAD + and the simultaneous reduction of a dye which leads to increased absorbance at OD = 450 nm, over 30 min. Reaction with buffer only serves as the negative control. N = 4–6. *p < 0.05. d ATP bioluminescent assay was performed to measure the level of intracellular ATP in heart tissues isolated from control and Tead1 icKO mice. N = 4–6. *p < 0.05. e-f CMs isolated at day 6 post the first tamoxifen injection from adult control Myh6-MerCreMer+; Tead1W/W or Myh6-MerCreMer+; Tead1F/F icKO mice. Subsequently cells were subjected to incubate with membrane-permeable, voltage-sensitive fluorescent probe TMRM (e, red) to quantify changes in mitochondrial membrane potential in live cells (f). *p < 0.05. g, h Similar to “e”, except that CMs treated with MitoSOX (green) to measure the production of superoxide by mitochondria. Cell nuclei were counter-stained with Hoechst 33258 (blue). The green fluorescence intensity of individual cells was quantified and plotted in “h”. *p < 0.05. i Using heart protein lysates from control or Tead1 icKO mice, Western blot was performed to determine the oxidized proteins that are marked by DNPH derivatization and probed by an anti-DNP antibody. j Quantification of the band intensity shown in “i” and normalized to respective loading control NDUFA9 (set to 1, red dashed line). N = 4. *p < 0.05. k Schematic diagram summarizing the major findings of this study. Transcription factor TEAD1 directly activates expression of nDNA-encoded mitochondrial genes that are essential for mitochondrial ETC activity, ATP production and cell survival in postmitotic CMs (upper panel). Conversely, Tead1 deletion represses mitochondrial genes especially complex I of ETC, leading to reduced ATP production and elevated oxidative stress. As a result, CMs deficient of Tead1 undergo necroptosis, which ultimately leads to DCM that can be partially rescued by the necroptosis blocker necrostatin-1 treatment (bottom panel).