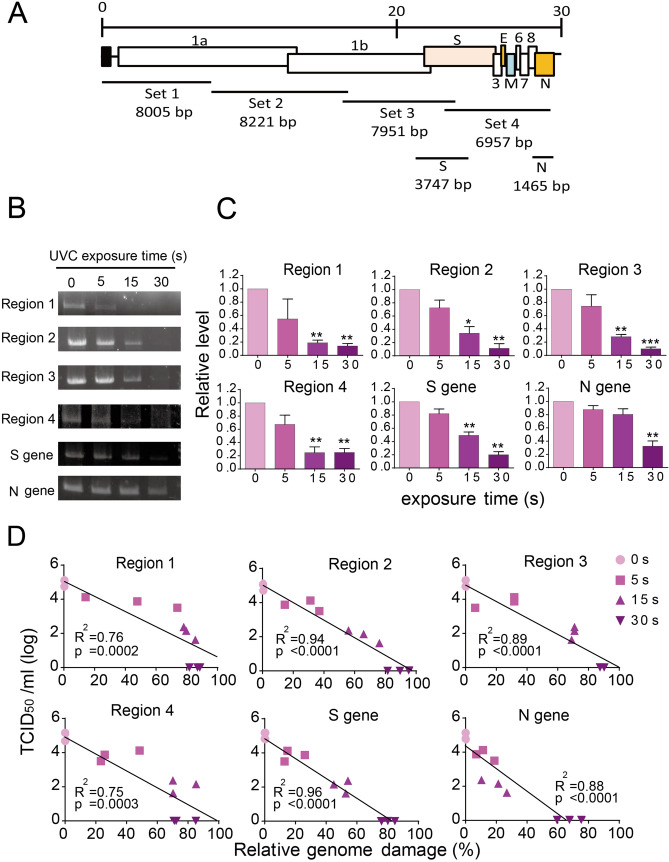
Figure 5.
Long reverse-transcription polymerase chain reaction (RT-PCR) to determine the effect of ultraviolet C (UVC) irradiation on the viral genome. (A) Schematic diagram of the six pairs of long RT-PCR primer sets. (B) Representative PCR products are visualized by agarose gel electrophoresis. Full-length gels are shown in Supplementary Fig. S2. SARS-CoV-2 (5 mL) with a titer of 5 × 104 50% tissue culture infective dose (TCID50/mL) was irradiated with a UVC light tube for increasing periods of time from 0 to 30 s. Viral RNA was reverse transcribed with the reverse primer for each region and then amplified with the corresponding primer sets. The amplified PCR products were used for 0.8% agarose gel electrophoresis and detected by ethidium bromide staining. (C) Band intensity is analyzed using CS Analyzer 4 software and quantitative results are shown in the bar diagram. Data in the plot represent the mean ± standard deviation (SD) of three replicates. Significance has been determined using one-way analysis of variance followed by Dunnett’s multiple comparisons test. The asterisk indicates a statistical difference (*p < 0.05; **p < 0.01; ***p < 0.001). (D) Linear regression analysis between relative viral genome damage and virus infectivity (derived from Fig. 2A). Relative genome damage is calculated as follows: (1-relative PCR fold change compared to control) × 100%.