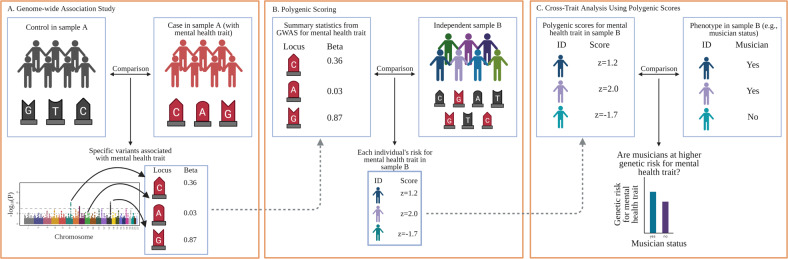
Fig. 4. Example of genome-wide association study (GWAS) and polygenic scores.
A GWAS are conducted by examining whether individual genetic loci (i.e., single-nucleotide polymorphisms, or SNPs, depicted with G, A, C, and T labels within a sample (or meta-analysis) differentiate cases from controls. The example is based on a dichotomous mental health trait (e.g., major depressive disorder diagnosis), but GWAS can be applied to other dichotomous and continuous phenotypes, such as trait anxiety, musician status, or hours of music practice. Importantly, rather than examining associations on a gene-by-gene basis, GWAS identify relevant genetic loci using SNPs from across the entire genome (typically depicted using a Manhattan plot, such as that displayed at the bottom of A). B After a GWAS has been conducted on a given trait, researchers can use the output to generate a polygenic score (sometimes called a polygenic risk score) in any new sample with genetic data by summing the GWAS effect sizes for each SNP allele present in a participant’s genome. An individual with a z = 2.0 would have many risk SNPs for that trait, whereas an individual with z = −2 would have much fewer risk SNPs. C Once a polygenic score is generated for all participants, it can be applied like any other variable in the new sample. In this example, researchers could examine whether musicians are at higher (or lower) genetic risk for a specific disorder. Other more complex analyses are also possible, such as examining how polygenic scores interact with existing predictors (e.g., trauma exposure) or polygenic scores for other traits to influence a phenotype or predict an intervention outcome. Created with BioRender.com.