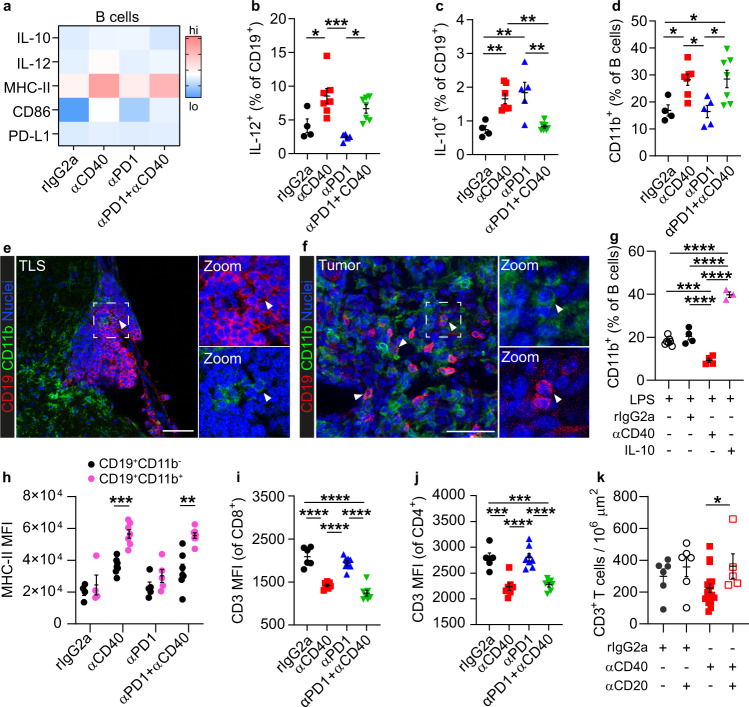
Fig. 6. Systemic delivery of αCD40 was associated with a CD11b+ regulatory phenotype of B cells.
All panels besides panel g show data from GL261 tumor-bearing mice. a Heatmap showing the expression levels of activation and immunosuppression markers on B cells in the brain, in the indicated treatment groups. b, c Quantification of b IL-12+ and c IL-10+ cells as a percentage of B cells in the brain, in the indicated treatment groups. b p(rIgG2a vs. αCD40) = 0.0175, p(αCD40 vs. αPD1) = 0.0005, p(αPD1 vs. αPD1+αCD40) = 0.0140. c p(rIgG2a vs. αCD40) = 0.0081, p(rIgG2a vs. αPD1) = 0.0031, p(αCD40 vs. αPD1+αCD40) = 0.0047, p(αPD1 vs. αPD1+αCD40) = 0.0018. d CD11b+ cells as a percentage of B cells in the brain, in the indicated treatment groups. p(rIgG2a vs. αCD40) = 0.0453; p(rIgG2a vs. αPD1+αCD40) = 0.0391¸ p(αCD40 vs. αPD1) = 0.0236; p(αPD1 vs. αPD1+αCD40) = 0.0201. a–d n(rIgG2a) = 4 mice, n(αCD40) = 7 mice, n(αPD1) = 5 mice, n(αPD1 + αCD40) = 7 mice. b–d One-way ANOVA with Tukey’s correction for multiple comparisons. e, f Immunofluorescent stainings showing CD11b+ B cells e in the TLS and f in the tumor area. Scale bars: 50 µm. g Quantification of CD11b+ cells as a percentage of wt murine splenic B cells after in vitro stimulation with the indicated agents. n(LPS) = 7 mice, n(LPS + rIgG2a) = 4 mice, n(LPS + αCD40) = 4 mice, n(LPS + IL-10) = 3 mice. One way ANOVA with Tukey’s correction for multiple comparisons. p(LPS vs. LPS+αCD40) = 0.0002, all other indicated p-values are p < 0.0001. h Mean fluorescence intensity (MFI) of MHC-II on CD11b+ vs CD11b− B cells in the brain, in the indicated treatment groups. n(rIgG2a) = 4 mice, n(αCD40) = 7 mice, n(αPD1) = 5 mice, n(αPD1 + αCD40) = 7 mice. p(αCD40) = 0.0002; p(αPD1+αCD40) = 0.0017. Multiple t-test with Sidak-Bonferroni correction. i, j MFI of CD3 on i CD8+ and j CD4+ T cells in the indicated treatment groups. One-way ANOVA with Tukey’s correction for multiple comparisons. i n(rIgG2a) = 6 mice, n(αCD40)&(αPD1 + αCD40) = 7 mice, n(αPD1) = 8 mice. All indicated p-values are p < 0.0001. j n(rIgG2a) = 5 mice, n(αCD40)&(αPD1 + αCD40) = 7 mice, n(αPD1) = 8 mice. p(rIgG2a vs. αCD40) = 0.0002, p(rIgG2a vs. αPD1+αCD40) = 0.0004. All indicated p-values are p < 0.0001. k Quantification of the number of CD3+ T cells in the tumor area in rIgG2a- or αCD40-treated mice with or without B cell depletion. n(rIgG2a)&(rIgG2a + αCD20) = 6 mice, n(αCD40) = 17 mice, n(αCD40 + αCD20) = 5 mice. p(αCD40 vs. αCD40-B) = 0.0442. One-way ANOVA with Tukey’s correction for multiple comparisons. b–d, i, j Black circle indicates rat IgG2a (rIgG2a), red square indicates agonistic CD40 antibodies (αCD40), blue triangle (pointing up) indicates PD1 blocking antibodies (αPD1), green triangle (pointing down) indicates αPD1 + αCD40. g Empty circle indicates lipopolysaccharide (LPS), black circle indicates LPS + rat IgG2a (rIgG2a), red square indicates LPS + agonistic CD40 antibodies (αCD40), pink triangle (pointing up) indicates LPS + interleukin 10 (IL-10). h Black circle indicates CD19+CD11b− B cells, pink circle indicated CD19+CD11b+. k Black circle indicates rIgG2a, empty circle indicates rIgG2a + αCD20, red square indicates αCD40, empty square indicates αCD40 + αCD20. For all graphs in this figure, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Bars: mean ± SEM. Source data are provided as a Source Data file.