Abstract
The accurate assay of cardiac troponin I (cTnI) is very important for acute myocardial infarction (AMI), it also can be employed as an effective index for screening serious patients in COVID-19 pandemic before fatal heart injury to reduce the mortality. A ratiometric sensing strategy was proposed based on electrochemiluminescent (ECL) signal of doxorubicin (Dox)-luminol or the electrochemical (EC) signal of methylene blue (MB) vs. referable EC signal of Dox. The bio-recognitive Tro4-aptamer ensures the high specificity of the sensor by affinity binding to catch cTnI, and the tetrahedral DNA (TDs) on Au/Ti3C2-MXene built an excellent sensing matrix. An in situ hybrid chain reaction (HCR) amplification greatly improved the sensitivity. The ratiometric sensing responses ECLDox-luminol/CurrentDox or CurrentMB/CurrentDox linearly regressed to cTnI concentration in the range of 0.1 fM-1 pM or 0.1 fM-500 fM with the limit of detection (LOD) as 0.04 fM or 0.1 fM, respectively. Served as the reference signal, CurrentDox reflected the variation of sensor, it is very effective to ensure the accuracy of detection to obviate the false results. The proposed biosensors show good specificity, sensitivity, reproducibility and stability, have been applied to determine cTnI in real samples with satisfactory results. They are worth looking forward to be used for screening serious patient of COVID-19 to reduce the mortality, especially in mobile cabin hospital.
Keywords: Cardiac troponin I, COVID-19 pandemic, Aptamer ratiometric sensor, Hybrid chain reaction, Tetrahedral DNA, Au/Ti3C2-MXene nanohybrid
1. Introduction
The global pandemic of COVID-19 pneumonia has lasted for more than one year (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). According to clinical researches, its high mortality directly relates to severe cases such as cardiac complications (Lukas et al., 2020; Shirazi et al., 2020). Cardiac troponin I (cTnI, high sensitivity troponin) is acknowledged as the most prominent biomarker for heart failure as in acute myocardial infarction (AMI), on account of its excellent specificity and sensitivity, also can provide long enough diagnostic window for myocardial injury (Han et al., 2016; Yang et al., 2019), its level in the blood mirrors the degree of myocardial injury, so the accuracy and sensitivity of cTnI measurements is of great significance for suspected heart patients (Zhu et al., 2019b). With a threshold of 0.04 ng/ml, a positive result of the cTnI concentration as 0.1 ng/ml clearly relates to microcirculation in acute myocardial infarction (Micro-AMI) patients (Fathil et al., 2015) and as high as 5–50 ng/ml for large AMI patients (Sun et al., 2019). Early, sensitive and accurate detection of AMI biomarkers can reduce mortality assuredly (Ji et al., 2019). The reports also revealed the cTnI rise in those COVID-19 patients (Guo et al., 2020; Huang et al., 2020), indicating that this index is important to identify severe cases from those mild patients, thus to take timely medical measures and therefore to reduce the mortality.
Until now, lots of researches focused on cTnI detection using immunosensors (Dong et al., 2020; Palanisamy et al., 2020; Yan et al., 2019; Yang et al., 2018; Ye et al., 2019; Zhu et al., 2019b), including electrochemiluminescent (ECL) ones and electrochemical (EC) ones with high selectivity and sensitivity (Dong et al., 2020; Jin et al., 2018; Labib et al., 2016; Li et al., 2018; Yang et al., 2018). However, those immunosensors were constructed by physically adsorbed or covalently immobilized antibodies on solid electrode surfaces in a random pattern, which may lead to in-homogenous distribution and random spatial orientation, resulting in a significant loss of antibody activity (Alves et al., 2012; Chen et al., 2014; Wang et al., 2017). This may eventually affect the reproducibility and sensitivity of the immunoassays and hamper their application. Recently, aptamer-based biosensors (aptasensors) have been constructed for cTnI assay as they offer many advantages to overcome the limitations of antibodies (Kitte et al., 2021; Sun et al., 2019). Aptamer is the single stranded DNA (ssDNA) or RNA produced in vitro with higher selectivity and affinity for proteid biomarker than antibody (Dhara and Mahapatra, 2020; Tao et al., 2020).
Herein the density and orientation of bio-recognitive probe on sensor surface is a core factor needed to be managed. Tetrahedral DNA (TDs) nanostructures, a novel “bottom-up” scaffolding strategy for probe loading, can perfectly regulate the density and orientation of probe packing. Also it provides the solution-phase-like environment to improve the probe-target recognition efficiency by effective elimination of nonspecific adsorption of non-targets (Chen et al., 2014, 2019; Huang et al., 2018; Pei et al., 2013, 2014). Thus it greatly promotes the application of biosensors (Lin et al., 2019; Sun et al., 2019; Zhu et al., 2019a).
The sensing signal amplification by natural enzymes-assistant (Zhang et al., 2016), nanozymatic ones (Dong et al., 2020; Wang et al., 2019; Ye et al., 2019; Zhou et al., 2014), and nucleotide ones (Yang et al., 2015; Zhang et al., 2020) are general measures for further improving the sensitivity. Among them, rolling circle amplification (RCA) (Liang et al., 2016) and hybridization chain reaction (HCR) (Dirks and Pierce, 2004; Zhu et al., 2019b) are widely used. Remarkably, HCR has been widely employed in bio-analysis (Bi et al., 2017; Li et al., 2018; Wei et al., 2018; Yang et al., 2019) as an isothermal enzyme-free amplification technique to trigger a tens to hundreds of repeated duplication (Zhang et al., 2020). Besides sensitivity and specificity, the accuracy is absolutely another important factor of biosensors. The ratiometry is relied on the ratio of two different signals rather than a single signal for quantitative analysis (Wu et al., 2017), which would improve the accuracy and reduce false results. A novel ratiometric strategy of ECL to EC was recently developed (Lin et al., 2019; Zhu et al., 2019a).
Ti3C2-MXene nanosheets are a novel family of 2D transition-metal carbonitride materials and have attracted considerable attention in various research fields (Fu et al., 2020; Kang et al., 2021; Wang et al., 2019; Wu et al., 2020; Zhang et al., 2020), on account of the excellent metallic conductivity, catalytic ability, good biocompatibility, hydrophilicity and high density of several functional groups (-F, –OH, –O) (Gao et al., 2017; Rakhi et al., 2015). Moreover, the Ti3C2-MXene nanosheets also possess strong reduction ability, making them quite popular in constructing Ti3C2-MXene/metal nanoparticle hybrids (Liu et al., 2019; Zhang et al., 2020; Zhang bet al., 2016). To date, Ti3C2-MXene-based EC, ECL, photoelectrochemical (PEC) and fluorescent sensing techniques have already been reported (Fu et al., 2020; Sun et al., 2020; Zhang et al., 2018, 2019). Doxorubicin (Dox), as an electroactive indicator, can be intercalated into double-stranded DNA (ds DNA) generated in HCR (Li et al., 2016, 2020; Yang et al., 2016), will act as a good signaler in this sensing approaching.
In this work, an elegant sensing strategy was achieved for sensitive and accurate assay of cTnI upon ECL/EC or EC/EC ratiometric response. By means of bi-function probe (BFP)/Tro4-aptamer/Au@Fe3O4 biocoalition, the combination of cTnI with Tro4-aptamer resulted in its disintegration to release the BFP, thereafter to hybridize with the capture probe on TDs/Au/Ti3C2-MXene sensing matrix. While the existence of two hairpins, an in situ HCR amplification will be initiated to form a double-stranded DNA (dsDNA) concatemer. Using a novel ECL luminophore (Dox-luminol complex, prepared by the cross-linking between Dox and luminol), amplified ECL signal is acquired because of their great number accommodation in dsDNA concatemer. The dsDNA of TDs could serve as the carrier of Dox, generating a stable internal reference EC signal to adjust potential variation of the sensor (ECL Dox-luminol/Current Dox). Another sensor with EC signal of methylene blue (MB, an electroactive indicator), enhanced in the presence of cTnI, also presented the sensing ability for cTnI (Current MB/Current Dox).
2. Experimental section
2.1. 1. Reagents, apparatus and materials preparation
These contents are described in Supplementary Material (S1–S6).
2.2. Fabrication of ratiometric aptasensors and the detection of cTnI
Prior to use, indium tin oxide (ITO) glasses were cleaned according to the previous report (Fu et al., 2020). 10 μL of 0.20% APTMS solution was then dropped on at 55 °C for 4 h to amino-functionalize its surface. After dried, 50 μL of AuNPs/Ti3C2-MXene suspension (l.5 mg/ml) was cast on this surface and kept at room temperature for 2 h, followed by rinsing with water and drying with N2. Then, 10 μL of TDs solution (1 μM) was incubated on at 4 °C for 12 h (TDs/Au/Ti3C2-MXene/ITO). TDs is anchored onto the surface of Au/Ti3C2-MXene/ITO via Au–S bonds from thiol groups at three vertices, emerged a pendant single DNA strand on last vertex. After that, 10 μL of Dox (2.0 mM) was placed on the sensing surface and incubated for 4 h at the room temperature to make them embedded into all edges of TDs.
After to incubate cTnI with BFP/Tro4-aptamer/Au@Fe3O4, equal amount of BFP was released. Driven out those magnetic components by external magnet, as-prepared TDs/Au/Ti3C2-MXene/ITO was immersed in it for 1.5 h at 37 °C. After washing, 10 μL of H1+H2 mixture (1 μM) was placed on the biosensor for incubation at 37 °C for 2.0 h, to fuel the HCR. Ultimately, the sensor was immersed in 10 μL of Dox-luminol complex as ECL indicators or 10 μL of 5 mM MB as EC indicator for 4 h at room temperature to adequately embed them into dsDNA. The sensing response was tested by ECL with a step voltage set up at −0.2 V for 2.7 s and 1.4 V for 0.3 s in turn in 0.2 M PBS of pH 7.4 containing 14 mM H2O2, and then immediately performed a negative SWV scan from −0.1 V to −0.8 V in 0.2 M PBS of pH 7.4 to obtain current signal of Dox, thus achieving the ratio of ECL Dox-Luminol/Current Dox as the output signal. The ratio of SWV Current MB/Current Dox was determined from −0.1 to −0.80 V in 0.2 M PBS (pH 7.4).
3. Results and discussion
3.1. 1. Principle of the ratiometric aptasensing strategy
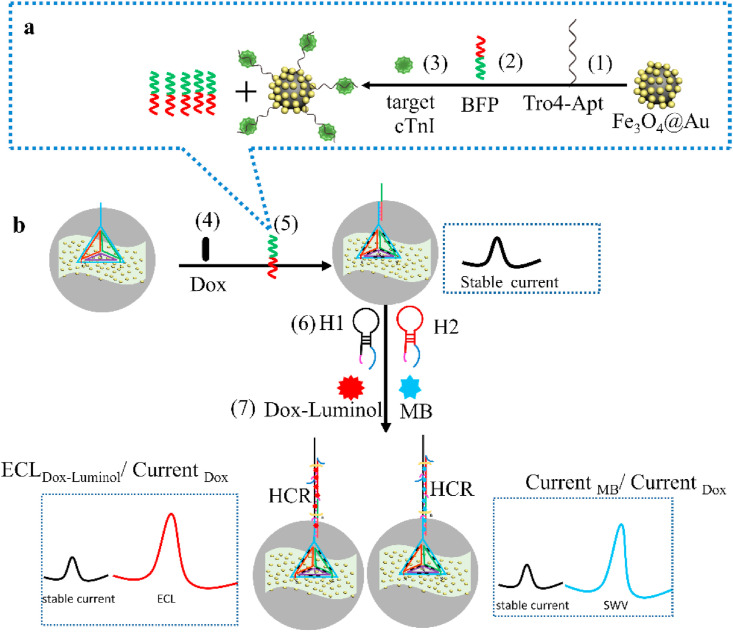
Scheme 1 illustrates the principle and construction of the ratiometric aptasensors. These TDs-supported probes greatly improve the target accessibility and minimize the non-specific adsorption on the sensor surface owing to its spatial structure, functional groups and dsDNA edge. A new bifunctional probe (BFP) (sequences in Table S1) was ingeniously designed and employed to rise specific hybridization and also to trigger HCR. In blank case, beside distinct SWV signal of Dox at −0.65 V, only very weak ECL signal of Dox-luminol or SWV signal of MB around −0.24 V were detected. Oppositely, the specific affinity reaction of cTnI with Tro4-aptamer liberated equal amount of BFP (as shown in Scheme 1a), initiated its hybridization with the capture DNA at vertex of TDs, meanwhile another part of BFP triggered the HCR with the coexistence of two hairpins (H1 and H2). Hence, the ECL probe (Dox-luminol) or EC indicator (MB) will be numerously bond on the long chain dsDNA concatemer, thereby generating a conspicuously amplified ECL or EC signal directly related to the cTnI concentration (as displayed in Scheme 1b). Dox, as an electrochemical indicator, would be intercalated into the grooves of the dsDNA on the edges of TDs to form Dox@TDs complex during the incubation on biosensor, to offer a stable EC signal as internal reference, to calibrate those differences between sensors or changes of microenvironment. Thus, the use of the dual-signal ratio of ECL Dox-Luminol/Current Dox or Current MB/Current Dox for the quantitative assay of the cTnI would be high accurate and reproducible. The successful synthesis of Dox-luminol was characterized by UV–Vis spectrum (see Fig. S1 and related discussion). The conjugation of BFP/Tro4-aptamer/Au@Fe3O4 biocoalition and Au@Fe3O4 nanocomposites were characterized by zeta potential, SEM, TEM, FT-IR, UV–Vis and EDS, see the details in SM, S4 and S5.
Scheme 1.
The illustration of (a) the specific target recognition and BFP release, and (b) ratiometric biosensing mechanism for cTnI.
3.2. 2. Characterization of TDs, HCR and BFP/aptamer composite
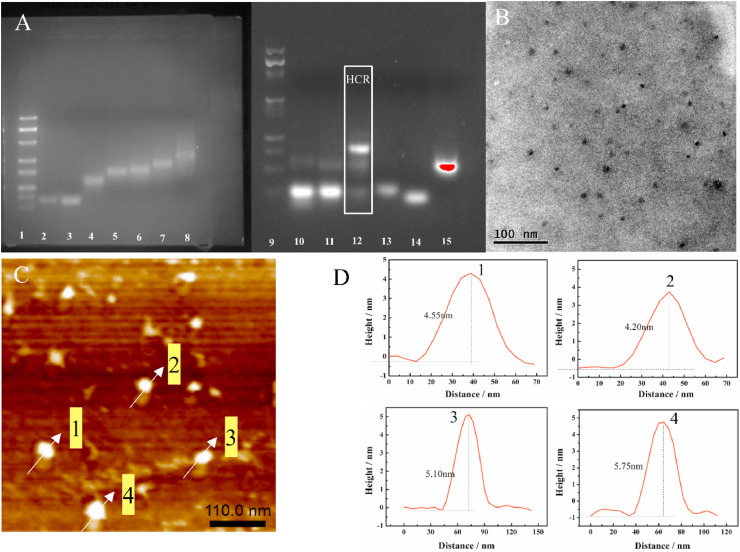
To confirm the successful assembly of TDs (the details of the preparation is presented in SM, S2, and Table S1 listed those sequences) and HCR, agarose gel electrophoresis was performed (Liu et al., 2019). As illustrated in Fig. 1 A, compared with the combinations of three strands (lane 6–7) and the single-strand or two strands DNA (lanes 2–5), TDs composed of four single strands (lane 8) showed slower mobility, in good consistent with its spatial complexity and increased mass (Lu et al., 2018). The only major clear band can be observed on the gel, suggesting its high yielding. The HCR process are initiated by cTnI in the presence of H1 and H2 as fuel strands triggered hairpin polymerization. Without cTnI, only one band could be found (Fig. 1A, lane 11), means no any hybridization happened. By introduction of cTnI, it would replace out BFP as the initiator of HCR, several bands were observed when H1 and H2 both existed in the solution (lane 12). At last, the HCR process would halt once two hairpins were exhausted. Compared to lane 13 and 14, lane 15 displayed more slowly migrating rate, indicating the successful hybridization of Tro4-aptamer and BFP.
Fig. 1.
(A) The gel electrophoretic image. 1, 100 bp DNA marker; 2, strand a; 3, strand b; 4, strand ab; 5, strand ad; 6, strand abc; 7, strand abd; 8, strand abcd (TDs); 9, 25 bp DNA marker; 10, H1; 11, after HCR in the absence of cTnI; 12, after HCR in the presence of cTnI; 13, Tro4-aptamer; 14, BFP; 15, Tro4-aptamer/BFP. (B) The TEM image of DNA tetrahedron. (C) The AFM image of TDs. (D) The height profiles of the TDs marked by the arrow in (C).
Transmission electron microscopy (TEM), and atomic force microscopy (AFM) were performed to further character the morphology and size of TDs (Lin et al., 2019). As shown in Fig. 1B, the approximate diameter (6.50 ± 0.8 nm) of TDs with uniform nanostructure estimated by TEM characterization was close to the calculated value (6.12 nm) (Lin et al., 2019; Zhu et al., 2019a). The AFM image exhibited TDs nanostructure (Fig. 1C) and Fig. 1D displayed the height range of TDs from 4.20 nm to 5.75 nm, close to theoretical value of the height and side length of TDs as 5.02 nm and 6.12 nm (Zhu et al., 2019a). All these results implied the correct formation of the TDs.
3.3. 3. Characterization of Ti3C2-MXene nanosheets and Au/Ti3C2-MXene nanocomposite
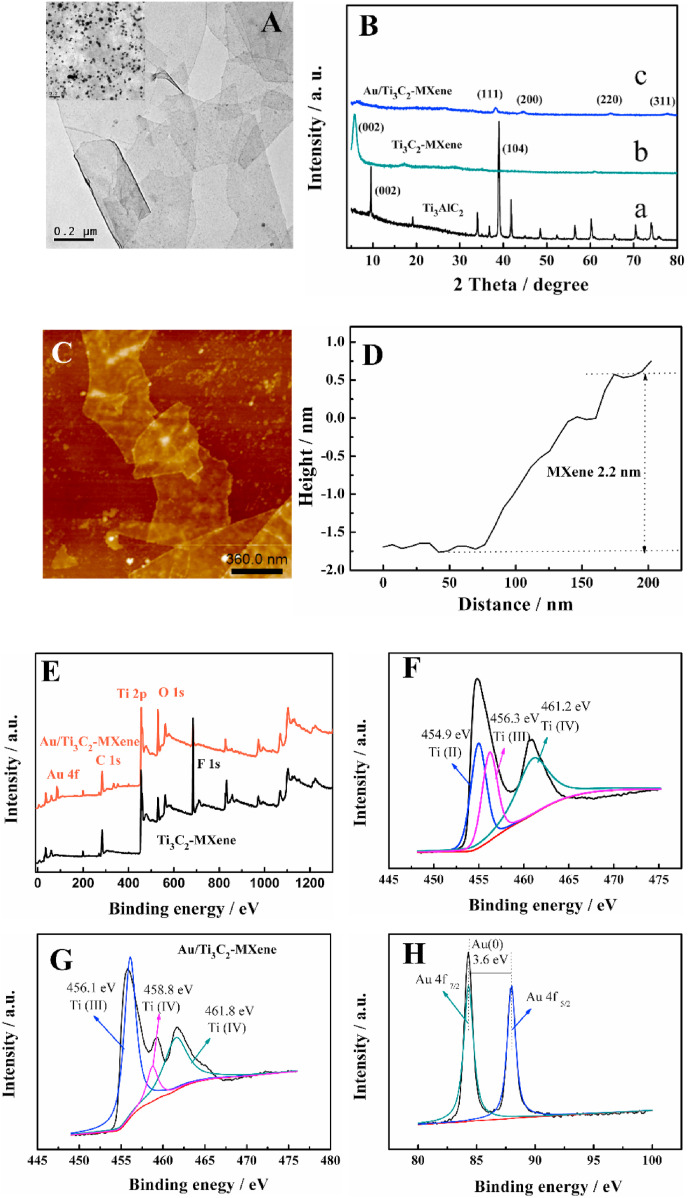
The successful synthesis of Ti3C2-MXenes was characterized by XRD, TEM and AFM. Fig. 2 A displays the TEM image of gauzy Ti3C2-MXene nanosheets after sonication exfoliating (Fu et al., 2020; Wu et al., 2020). The TEM image also clearly shows that the AuNPs with <30 nm of diameter were decorated on the Ti3C2-MXene nanosheets (inserted in Fig. 2A). The XRD curve in Fig. 2B indicates the disappearance of the characteristic peak of Ti3AlC2 (curve a) at 39° and the shift of (002) peak to lower angle, demonstrating the transformation of Ti3AlC2 to Ti3C2-MXene (curve b), in good agreement with a previous report (Liu et al., 2019). The morphology and thickness of Ti3C2-MXene nanosheet was analyzed by AFM (Fig. 2C and D) as about 2.2 nm, indicating that few or single layer Ti3C2-MXene nanosheet was formed by sonication. After homogeneous self-reduction of AuCl4 − ions, four characteristic diffraction peaks of AuNPs appeared on the XRD pattern (Fig. 2B, curve c). In addition, compared with the (002) peak intensity of Ti3C2-MXene nanosheet, the weak peak of Au/Ti3C2-MXene could be attributed to partial oxidation of Ti3C2-MXene nanosheets during the self-reduction reaction of AuCl4 − ions (Zhang et al., 2020). These results all support the successful synthesis of Au/Ti3C2-MXene nanocomposite.
Fig. 2.
(A) The TEM image of Ti3C2-MXene (inserted is the TEM image of Au/Ti3C2-MXene). (B) The XRD pattern of Ti3AlC2, Ti3C2-MXene and Au/Ti3C2-MXene. (C) The AFM image of Ti3C2-MXene and (D) the thickness of Ti3C2-MXene. (E) The XPS profiles of the Au/Ti3C2-MXene, (F) The elemental valences of Ti in Ti3C2-MXene, The elemental valence of (G) Ti and (H) Au in Au/Ti3C2-MXene.
Raman, infrared, UV–Vis and energy dispersive X-ray (EDS, Figs. S5A-S5D) spectra characterized the successful synthesis of Ti3C2-MXene nanosheets and the decoration of AuNPs on it. To illuminate the self-reduction mechanism of AuCl4 - by Ti3C2-MXene, XPS analysis (Fig. 2E–H) was performed. The wide peak from 450 to 460 eV in Ti3C2-MXene implied the possible existence of low valence Ti species, such as Ti2+ at ~454.9 eV (Zou et al., 2016), acted as the origin of Au anion reduction. The representative Au 3d5/2 and Au 3d7/2 peaks in the Au/Ti3C2-MXene composite located at ~87.3 and ~83.7 eV with the binding energy gap of ~3.6 eV, suggesting the successful formation of Au (0) (Zhang et al., 2020). The catalytic effect of Ti3C2-MXene nanosheet on ECL of luminol was investigated, presenting indeed enhancement (S7 in SM). What's more, the proposed Ti3C2-MXene and Au/Ti3C2-MXene modified GCE displayed excellent electron transfer ability (see Fig. S7A).
3.4. Characterization of the preparation and the feasibility of the aptasensors
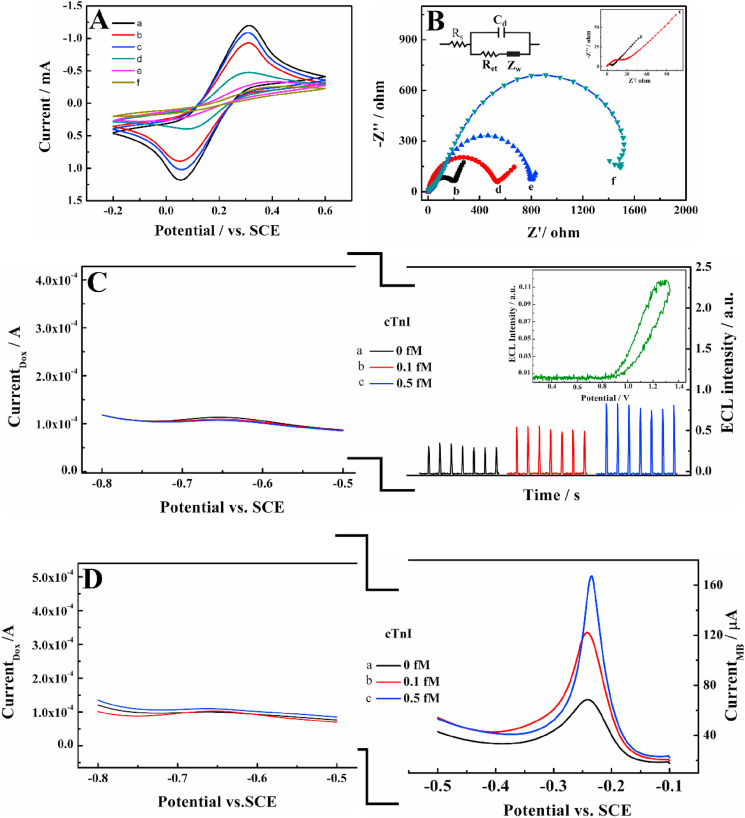
In 0.1 M KCl solution containing 5 mM Fe(CN)6 3−/4-, the cyclic voltammetric (CV) curves of different electrodes are recorded (in Fig. 3 A). A couple of reversible redox peaks around 0.3 V was observed on bare ITO (curve a). When APTMS was grafted on it, peak currents decreased due to its poor conductivity (curve b). After Au/Ti3C2-MXene nanosheets were attached on by Au–N bond, larger peak currents appeared, because of the excellent conductivity of AuNPs and Ti3C2-MXene nanosheets (curve c). According to Randles-Sevcik equation, the electrochemical active area of Au/Ti3C2-MXene/ITO can be calculated to be 0.65 cm2 (see detailed discussion in S8, SM). Then, TDs probes were assembled on, inducing distinctly decrease in peak current because of the electrostatic repulsive interaction toward Fe(CN)6 3−/4- (curve d). Here, the quantity of the TDs was measured as 3.72 × 1012 (S9 in SM), given enough binding sites for the immobilization of BFP. Finally, along with the assembly of BFP and HCR on the electrode surface, the CV peak currents successively minimized (curves e-f), indicating the interfacial steric hindrance of those macro biomolecules.
Fig. 3.
The (A) CV and (B) EIS of (a) ITO, (b) APTMS/ITO, (c) Au/Ti3C2-MXene/ITO, (d) TDs/Au/Ti3C2-MXene/ITO, (e) BFP/TDs/Au/Ti3C2-MXene/ITO and (f) after HCR. Inset of B is the Randles equivalent circuit. (C) The ECL/EC signals of biosensor before and after use. Inset of (C) is the potential-intensity curve of luminol ECL. (D) The EC signals of MB and Dox in the absence and presence of cTnI.
Electrochemical impedance spectroscopy (EIS) is powerful to evaluate the electron transfer resistance (R et) of sensor substrate, reflecting the surface alteration. As displayed in Fig. 3B, all of the Nyquist plots were fitted to the Randles equivalent circuit (Fig. 3B, inset), where R s, C d, Z W, R et express their conventional meaning. Compared to small R et of bare ITO (inset curve a), the coverage of APTMS on its surface induced a large R et due to the formation of this insulation layer (curve b). Afterward, the anchored Au/Ti3C2-MXene largely decreased the R et, attributing to its excellent metallic conductivity and thereby improvement of electron transfer (inset curve c). However, the R et augmented consecutively with the sequential assembly of TDs, BFP and HCR (curves d to f), which can be explained by the success of progressive assembly of cTnI biosensor, hindering the electron transfer with Fe(CN)6 3−/4-, consistent with that observed in CV.
To further validate the feasibility of the ECL/EC ratiometric strategy, the alteration of ECL intensities and SWV signals were recorded in conditions of whether is there a target. As displayed in Fig. 3C, in the absence of cTnI, occupied by hairpins, little amount of Dox-luminol can interact onto sensor surface, giving only weak ECL signal. Thereafter, along with the increase of cTnI concentration, the ECL signal was significantly enhanced. A significant SWV signal of Dox intercalated in TDs at −0.65 V shows small distinction upon different sensor or microenvironment, implying that the EC signal of Dox was uncorrelated to the target. Hence, the ratio of ECL Dox-Luminol/Current Dox could eliminate the variation of sensors to improve the accuracy of detection. The ECL emission of luminol raised at about 1.3 V (Fig. 3C, inset), without cross action with the oxidation potential of Dox. So the stable EC signal of Dox can be employed as the reference signal. Likewise, the feasibility of the EC/EC ratiometric strategy was also confirmed in Fig. 3D, similarly, cTnI concentration can be calibrated based on the ratio of responses (Current MB/Current Dox).
3.5. 5. cTnI aptasensing
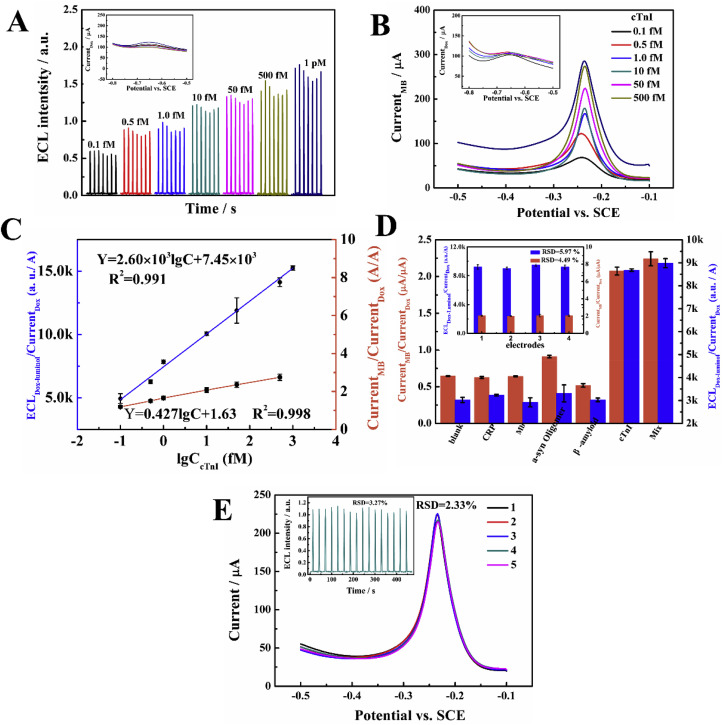
The analytical performances of the proposed ratiometric biosensors for cTnI were investigated under the optimal conditions (see S10 in SM): 0.20% APTMS, 1.5 mg/ml Au/Ti3C2-MXene suspension, 14 mM H2O2, 240 min for the intercalation of Dox-Luminol or MB respectively. In Fig. 4 A, as the sensing signal, the ECL intensity of luminol gradually increased along with the cTnI concentration from 0.1 fM to 1 pM, while internal reference signal of Dox current at about −0.65 V was stable. A good linear correlation between the ratio value (ECL Dox-Luminol/Current Dox) as the output vs. the logarithm of cTnI concentration was obtained (Fig. 4C): ECL Dox-Luminol/Current Dox (a.u./A) = 2.60 × 103lgC+7.45 × 103, R2 = 0.991, with a limit of detection (LOD) as 0.04 fM (0.96 fg/ml) (S/N = 3). Fig. 4B displays the response of the sensor in EC/EC mode, the ratio value (Current MB/Current Dox) linearly depended on the logarithm of cTnI concentration within the range of 0.1 fM to 500 fM (Fig. 4C). The linear equation is Current MB/Current Dox = 0.427lgC+1.63 (R2 = 0.998, with an LOD as 0.1 fM, 2.4 fg/ml, S/N = 3). At the same time, the comparison of luminol ECL signal towards TDs-based biosensor and ssDNA revealed its better suitability for cTnI assay (shown in Fig. S10, SM). Another control experiment with poly-T strand to work instead of aptamer was carried out, it presented a background ECL emission in roughly the same as blank aptasensor, but nothing changed anymore in sensing process (column “g”, shown in Fig. S10), demonstrates the exclusive necessity of Tro4-aptamer for sensor response, verdicting that the binding of the Tro4-aptamer with the target triggered the HCR amplification process (see detailed discussion in S11, SM). In summary, compared to reported bioassays of cTnI (Table S2), these two sensors displayed higher sensitivity.
Fig. 4.
(A) The ECL and SWV curves (inset) upon cTnI concentration. (B) The SWV signals of MB and Dox upon cTnI concentration. (C) The calibration curve of ECLDox-Luminol/CurrentDox and CurrentMB/CurrentDox vs. lgCcTnI. (D) The selectivity of ECL/EC and EC/EC ratiometric biosensors (10 fM cTnI vs. 500 fM nontarget proteins), the inset is the reproducibility of developed ECL/EC and EC/EC biosensors for 10 fM cTnI with four parallel sensors, Error bar = Standard deviation (SD, n = 3). (E) The stability of proposed EC/EC and ECL/EC biosensor (inset) for 10 fM cTnI under 5 cycles or 16 cycles of successive scans.
The specificity of the sensors was evaluated toward cTnI against four potential interferences including C-reactive protein (CRP), myoglobin (Mb), β-amyloid, and α-syn oligomer. As can be seen from Fig. 4D, for beyond 50 multiple higher concentration of those matters than cTnI, they do not disturb the detection of cTnI. Using four parallel sensors to detect 10 fM cTnI, the relative standard deviation (RSD, i.e. coefficient of variation) of the ECL Dox-Luminol/Current Dox and Current MB/Current Dox responses are 5.97% and 4.49%, respectively (inset of Fig. 4D). Also the stability of the constructed biosensors was assessed by incubation with 10 fM cTnI under 16 cycles and 5 cycles of consecutive scanning, respectively. As displayed in Fig. 4E, the RSD of ECL response (inset of Fig. 4E) was 3.27%, and RSD of EC signal was 2.33%, respectively, suggesting that the designed biosensors were stable.
3.6. 6. Real sample analysis
To evaluate the practicability of the developed biosensors, the cTnI content in 3 normal human serum samples (supplied by the Second Affiliated Hospital of Soochow University, stored in −20 °C) were tested after the dilution of 100 times with 0.2 M PBS (pH 7.4), and also the recoveries were tested using standard addition method. As listed in Table 1 , the good recoveries (90.6%–99.4%) of ECL/EC sensor and (93.3%–101.4%) of EC/EC sensor are achieved. The good recoveries suggested that the ratiometric biosensors could work well in complex samples. Also other 4 samples from the AMI patient were examined parallelly with sensors and ELISA kit (Table 1), got a small error less than 6.0%, indicating good accuracy of the proposed biosensors.
Table 1.
| Sample | cTnI found by ECL/EC (fM) | cTnI added fM) | detected by ECL/EC sensor (fM) | recovery (%) | detected by EC/EC sensor (fM) | recovery (%) |
|---|---|---|---|---|---|---|
| Healthy1 | 1.21 ± 0.01 | 3.00 | 4.19 ± 0.38 | 99.4 | 4.25 ± 0.19 | 101.4 |
| Healthy2 | 0.83 ± 0.01 | 5.00 | 5.36 ± 0.07 | 90.6 | 5.49 ± 0.16 | 93.3 |
| Healthy3 | 0.65 ± 0.01 | 7.00 | 7.20 ± 1.21 | 93.6 | 7.30 ± 0.45 | 95.0 |
| Sample |
ELISA (fM) |
detected by ECL/EC sensor (fM) |
RSD (%) |
detected by EC/EC sensor (fM) |
RSD (%) |
|
| Patient 1 | 1.25 | 1.25 ± 0.03 | 2.76 | 1.22 ± 0.07 | 5.32 | |
| Patient 2 | 15.0 | 14.7 ± 0.85 | 5.80 | 15.2 ± 0.73 | 4.80 | |
| Patient 3 | 2.53 × 103 | 2.42 × 103±83.5 | 3.45 | 2.45 × 103±122 | 5.00 | |
| Patient 4 | 2.20 × 103 | 2.13 × 103±59.5 | 2.79 | 2.19 × 103±125 | 5.70 | |
Mean value ± SD of three independent experiments (n = 3), RSD (n = 3).
Mean value ± SD of three independent experiments (n = 3), RSD (n = 3).
4. Conclusion
In this research, fascinating ratiometric ECL/EC and EC/EC aptasensors were constructed by integrating aptamer-based molecular recognition with the ECL or EC signal transduction and dual amplification for cTnI detection. As we all know, people with one or more high risk factors such as hypertension, hyperlipidemia, thrombus etc, need early and regular medical inspection to manage their daily life although who has not yet shown any AMI symptoms like pectoralgia; for people who have had myocardial infarction, it is indispensable to monitor the condition more frequently. While the Electrocardiogram (ECG) is almost impossible for home use, the assay of AMI biomarkers as cTnI in peripheral blood is a more preferable way. Take the lead to design cTnI sensor with two-channel signal outputs in this work, more accurate detection is expectable with lower-level of false result, because the false results in AMI diagnosis is a life-threatening factor. It will help the improvement of reliability of clinical assay, thereafter to promote the comprehensive benefits of medical therapy. Meanwhile, beside ELISA, with higher sensitivity attributed to the ability of multi-amplification in this sensor, we path a new way for clinical staffs to get earlier precaution of AMI to save more lives, especially in the case of global pandemic of COVID-19, which would give hints to deal with the patients to block-up the turning from mild to severe. Coupled with portable ECL/EC workstation (Tan et al., 2021; Tu et al., 2020), the aptasensors are very probable to be employed for screening the symptom of heart injury in those COVID-19 patients, especially the use in mobile cabin hospital.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (21675115, 21375091).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113482.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alves N.J., Kiziltepe T., Bilgicer B. Langmuir. 2012;28(25):9640–9648. doi: 10.1021/la301887s. [DOI] [PubMed] [Google Scholar]
- Bi S., Yue S.Z., Zhang S.S. Chem. Soc. Rev. 2017;46(14):4281–4298. doi: 10.1039/c7cs00055c. [DOI] [PubMed] [Google Scholar]
- Chen X., Huang J., Zhang S., Mo F., Su S.S., Li Y., Fang L.C., Deng J., Huang H., Luo Z.X., Zheng J.S. ACS Appl. Mater. Interfaces. 2019;11(4):3745–3752. doi: 10.1021/acsami.8b20144. [DOI] [PubMed] [Google Scholar]
- Chen X.Q., Zhou G.B., Song P., Wang J.J., Gao J.M., Lu J.X., Fan C.H., Zuo X.L. Anal. Chem. 2014;86(15):7337–7342. doi: 10.1021/ac500054x. [DOI] [PubMed] [Google Scholar]
- Dhara K., Mahapatra D.R. Microchem. J. 2020;156:104857–104881. [Google Scholar]
- Dirks R.M., Pierce N.A. PANS (Pest. Artic. News Summ.) 2004;101(43):15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Cao L.L., Tan Z.L., Liu Q., Zhou J., Zhao P.P., Wang P., Li Y.Y., Ma W.S., Dong Y.H. ACS Appl. Bio Mater. 2020;3(1):377–384. doi: 10.1021/acsabm.9b00863. [DOI] [PubMed] [Google Scholar]
- Fathil M.F.M., Md Arshad M.K., Gopinath S.C.B., Hashim U., Adzhri R., Ayub R.M., Ruslinda A.R., Nuzaihan M.N.M., Azman A.H., Zaki M., Tang T.H. Biosens. Bioelectron. 2015;70:209–220. doi: 10.1016/j.bios.2015.03.037. [DOI] [PubMed] [Google Scholar]
- Fu Y.M., Ding F., Chen J.H., Liu M.Y., Zhang X.H., Du C.C., Si S.H. Chem. Commun. 2020;56(43):5799–5802. doi: 10.1039/d0cc01835j. [DOI] [PubMed] [Google Scholar]
- Gao G.P., O'Mullane A.P., Du A.J. ACS Catal. 2017;7(1):494–500. [Google Scholar]
- Guo T., Fan Y.Z., Chen M., Wu X.Y., Zhang L., He T., Wang H.R., Wan J., Wang X.H., Lu Z.B. JAMA Cardiol. 2020;5(7):811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Li S.H., Peng Z.L., Othman A.M., Leblanc R. ACS Sens. 2016;1(2):106–114. [Google Scholar]
- Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., Zhang L., Fan G.H., Xu J.Y., Gu X.Y., Cheng Z.S., Yu T., Xia J.A., Wei Y., Wu W.J., Xie X.L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J.G., Wang G.F., Jiang R.M., Gao Z.C., Jin Q., Wang J.W., Cao B. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.R., He N.Y., Li Z.Y. Biosens. Bioelectron. 2018;109:27–34. doi: 10.1016/j.bios.2018.02.053. [DOI] [PubMed] [Google Scholar]
- Ji J.J., Lu W.B., Zhu Y., Jin H., Yao Y.Y., Zhang H.D., Zhao Y.J. ACS Sens. 2019;4(5):1384–1390. doi: 10.1021/acssensors.9b00352. [DOI] [PubMed] [Google Scholar]
- Jin Y.C., Kang Q., Guo X.L., Zhang B., Shen D.Z., Zou G.Z. Anal. Chem. 2018;90(21):12930–12936. doi: 10.1021/acs.analchem.8b03554. [DOI] [PubMed] [Google Scholar]
- Kang Z.M., Khan M.A., Gong Y.M., Javed R.D., Xu Y., Ye D.X., Zhao H.B., Zhang J.J. J. Mater. Chem. 2021:6089–6108. [Google Scholar]
- Kitte S.A., Tafese T., Xu C., Saqib M., Li H.J., Jin Y.D. Talanta. 2021;221:121674–121681. doi: 10.1016/j.talanta.2020.121674. [DOI] [PubMed] [Google Scholar]
- Labib M., Sargent E.H., Kelley S.O. Chem. Rev. 2016;116(16):9001–9090. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- Li D.D., Xu Y.X., Fan L., Shen B., Ding X.J., Yuan R., Li X.M., Chen W.X. Biosens. Bioelectron. 2020;148:111826–111833. doi: 10.1016/j.bios.2019.111826. [DOI] [PubMed] [Google Scholar]
- Li J.Y., Liu S.Y., Sun L.Q., Li W., Zhang S.Y., Yang S., Li J., Yang H.H. J. Am. Chem. Soc. 2018;140(48):16589–16595. doi: 10.1021/jacs.8b08442. [DOI] [PubMed] [Google Scholar]
- Li W.S., Yang X.H., He L.L., Wang K.M., Wang Q., Huang J., Liu J.B., Wu B., Xu C.C. ACS Appl. Mater. Interfaces. 2016;8(39):25733–25740. doi: 10.1021/acsami.6b08210. [DOI] [PubMed] [Google Scholar]
- Li Y.Y., Zhu W.J., Kang Q., Yang L., Zhang Y., Wang Y.G., Wei Q. ACS Appl. Mater. Interfaces. 2018;10(45):38791–38798. doi: 10.1021/acsami.8b14908. [DOI] [PubMed] [Google Scholar]
- Liang W.B., Fan C.C., Zhuo Y., Zheng Y.N., Xiong C.Y., Chai Y.Q., Yuan R. Anal. Chem. 2016;88(9):4940–4948. doi: 10.1021/acs.analchem.6b00878. [DOI] [PubMed] [Google Scholar]
- Lin Y., Jia J.P., Yang R., Chen D.Z., Wang J., Luo F., Guo L.H., Qiu B., Lin Z.Y. Anal. Chem. 2019;91(5):3717–3724. doi: 10.1021/acs.analchem.9b00013. [DOI] [PubMed] [Google Scholar]
- Liu D., Zhang G., Ji Q.H., Zhang Y.Y., Li J.H. ACS Appl. Mater. Interfaces. 2019;11(29):25758–25765. doi: 10.1021/acsami.9b02511. [DOI] [PubMed] [Google Scholar]
- Liu Z., Lei S., Zou L.N., Li G.P., Xu L.L., Ye B.X. Biosens. Bioelectron. 2019;131:113–118. doi: 10.1016/j.bios.2019.02.020. [DOI] [PubMed] [Google Scholar]
- Lu H.J., Pan J.B., Wang Y.Z., Ji S.Y., Zhao W., Luo X.L., Xu J.J., Chen H.Y. Anal. Chem. 2018;90(17):10434–10441. doi: 10.1021/acs.analchem.8b02347. [DOI] [PubMed] [Google Scholar]
- Lukas H., Xu C.H., Yu Y., Gao W. ACS Nano. 2020;14(12):16180–16193. doi: 10.1021/acsnano.0c08494. [DOI] [PubMed] [Google Scholar]
- Palanisamy S., Senthil Raja D., Subramani B., Wu T.H., Wang Y.M. ACS Appl. Mater. Interfaces. 2020;12(29):32468–32476. doi: 10.1021/acsami.0c09086. [DOI] [PubMed] [Google Scholar]
- Pei H., Zuo X.L., Pan D., Shi J.Y., Huang Q., Fan C.H. NPG Asia Mater. 2013;5(6):51–60. [Google Scholar]
- Pei H., Zuo X.L., Zhu D., Huang Q., Fan C.H. Acc. Chem. Res. 2014;47(2):550–559. doi: 10.1021/ar400195t. [DOI] [PubMed] [Google Scholar]
- Rakhi R.B., Ahmed B., Hedhili M.N., Anjum D.H., Alshareef H.N. Chem. Mater. 2015;27(15):5314–5323. [Google Scholar]
- Shirazi S., Mami S., Mohtadi N., Ghaysouri A., Tavan H., Nazari A., Kokhazadeh T., Mollazadeh R. Future Cardiol. 2020;17(1):113–118. doi: 10.2217/fca-2020-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.P., Luo Z.B., Lu J., Zhang S.S., Che T., Chen Z.G., Zhang L.Y. Biosens. Bioelectron. 2019;134:49–56. doi: 10.1016/j.bios.2019.03.049. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zhang Y.M., Zhang H.X., Liu M.L., Liu Y. Anal. Chem. 2020;92(15):10668–10676. doi: 10.1021/acs.analchem.0c01776. [DOI] [PubMed] [Google Scholar]
- Tan R., Shen Y., Li D.N., Yang Y., Tu Y.F. Electroanalysis. 2021;33:1–10. [Google Scholar]
- Tao X.Q., Wang X., Liu B.W., Liu J.W. Biosens. Bioelectron. 2020;168:112537–112551. doi: 10.1016/j.bios.2020.112537. [DOI] [PubMed] [Google Scholar]
- Tu Y.F., Shen Y., Liu Y.H., Tan R. Chinese patent. 2020;4 [Google Scholar]
- Wang H., Li H., Huang Y., Xiong M.H., Wang F., Li C. Biosens. Bioelectron. 2019;142:111531–111538. doi: 10.1016/j.bios.2019.111531. [DOI] [PubMed] [Google Scholar]
- Wang J.P., Leong M.C., Leong E.Z.W., Kuan W.S., Leong D.T. Anal. Chem. 2017;89(12):6900–6906. doi: 10.1021/acs.analchem.7b01508. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Zhao Y.Y., Wang M.M., Li H.J., Saqib M., Ge C.H., Zhang X.D., Jin Y.D. Anal. Chem. 2019;91(4):3048–3054. doi: 10.1021/acs.analchem.8b05443. [DOI] [PubMed] [Google Scholar]
- Wei J., Chang W.D., Qileng A., Liu W.P., Zhang Y., Rong S.Y., Lei H.T., Liu Y.J. Anal. Chem. 2018;90(15):9606–9613. doi: 10.1021/acs.analchem.8b02546. [DOI] [PubMed] [Google Scholar]
- Wu L., Ding F., Yin W.M., Ma J., Wang B.R., Nie A., Han H.Y. Anal. Chem. 2017;89(14):7578–7585. doi: 10.1021/acs.analchem.7b01399. [DOI] [PubMed] [Google Scholar]
- Wu Q., Li N.B., Wang Y., Xu Y.C., Wu J.D., Jia G.R., Ji F.J., Fang X.D., Chen F.F., Cui X.Q. Anal. Chem. 2020;92(4):3354–3360. doi: 10.1021/acs.analchem.9b05372. [DOI] [PubMed] [Google Scholar]
- Yan M.X., Ye J., Zhu Q.J., Zhu L.P., Huang J.S., Yang X.R. Anal. Chem. 2019;91(15):10156–10163. doi: 10.1021/acs.analchem.9b02169. [DOI] [PubMed] [Google Scholar]
- Yang G.X., Zhuang H.S., Chen H.Y., Ping X., Bu D. Sens. Actuators, B. 2015;214:152–158. [Google Scholar]
- Yang J., Shen H.W., Zhang X., Tao Y.Y., Xiang H., Xie G.M. Biosens. Bioelectron. 2016;77:1119–1125. doi: 10.1016/j.bios.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Yang R., Li F., Zhang W.C., Shen W., Yang D., Bian Z.P., Cui H. Anal. Chem. 2019;91(20):13006–13013. doi: 10.1021/acs.analchem.9b03066. [DOI] [PubMed] [Google Scholar]
- Yang W., Peng Q.L., Guo Z., Wu H.P., Ding S.J., Chen Y.J., Zhao M. Biosens. Bioelectron. 2019;142:111548–111556. doi: 10.1016/j.bios.2019.111548. [DOI] [PubMed] [Google Scholar]
- Yang X., Yu Y.Q., Peng L.Z., Lei Y.M., Chai Y.Q., Yuan R., Zhuo Y. Anal. Chem. 2018;90(6):3995–4002. doi: 10.1021/acs.analchem.7b05137. [DOI] [PubMed] [Google Scholar]
- Ye J., Zhu L.P., Yan M.X., Zhu Q.J., Lu Q.Q., Huang J.S., Cui H., Yang X.R. Anal. Chem. 2019;91(2):1524–1531. doi: 10.1021/acs.analchem.8b04640. [DOI] [PubMed] [Google Scholar]
- Zhang C.Y., Chen J., Sun R., Huang Z.J., Luo Z.W., Zhou C., Wu M.F., Duan Y.X., Li Y.X. ACS Sens. 2020;5(10):2977–3000. doi: 10.1021/acssensors.0c01453. [DOI] [PubMed] [Google Scholar]
- Zhang H.X., Wang Z.H., Wang F., Zhang Y.M., Wang H.Y., Liu Y. Anal. Chem. 2020;92(7):5546–5553. doi: 10.1021/acs.analchem.0c00469. [DOI] [PubMed] [Google Scholar]
- Zhang N., Ma Z.Y., Ruan Y.F., Zhao W.W., Xu J.J., Chen H.Y. Anal. Chem. 2016;88(4):1990–1994. doi: 10.1021/acs.analchem.5b04579. [DOI] [PubMed] [Google Scholar]
- Zhang Q.X., Wang F., Zhang H.X., Zhang Y.Y., Liu M.L., Liu Y. Anal. Chem. 2018;90(21):12737–12744. doi: 10.1021/acs.analchem.8b03083. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Jiang X.T., Zhang J.J., Zhang H., Li Y.C. Biosens. Bioelectron. 2019;130:315–321. doi: 10.1016/j.bios.2019.01.043. [DOI] [PubMed] [Google Scholar]
- Zhang Z.W., Li H.N., Zou G.D., Fernandez C., Liu B.Z., Zhang Q.R., Hu J., Peng Q.M. ACS Sustain. Chem. Eng. 2016;4(12):6763–6771. [Google Scholar]
- Zhou Y., Zhuo Y., Liao N., Chai Y.Q., Yuan R. Chem. Commun. 2014;50(93):14627–14630. doi: 10.1039/c4cc05269b. [DOI] [PubMed] [Google Scholar]
- Zhu L.P., Ye J., Wang S., Yan M.X., Zhu Q.J., Huang J.S., Yang X.R. Chem. Commun. 2019;55(77):11551–11554. doi: 10.1039/c9cc05592d. [DOI] [PubMed] [Google Scholar]
- Zhu L.P., Ye J., Yan M.X., Zhu Q.J., Wang S., Huang J.S., Yang X.R. ACS Sens. 2019;4(10):2778–2785. doi: 10.1021/acssensors.9b01369. [DOI] [PubMed] [Google Scholar]
- Zou G.D., Zhang Z.W., Guo J.X., Liu B.Z., Zhang Q.R., Fernandez C., Peng Q.M. ACS Appl. Mater. Interfaces. 2016;8(34):22280–22286. doi: 10.1021/acsami.6b08089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.