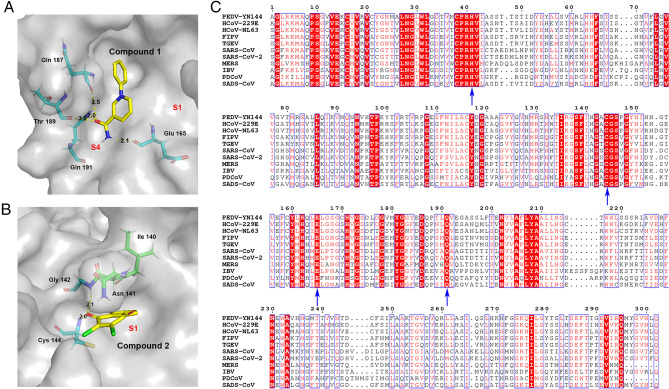
Fig. 5.
In silico molecular docking of compounds 1 and 2 with PEDV 3CLpro. A Docked conformation of compound 1 with PEDV 3CLpro. The protease is shown via a charge-potential surface. The possible hydrogen bond interactions are presented as yellow dashed lines, and the bond distances are shown. The S1 and S4 pockets are labelled. B Docked conformation of compound 2 with PEDV 3CLpro. The compounds and interactions between compounds and binding sites are represented as in A. C Sequence alignment of various coronavirus 3CLpros. The amino acid sequence information was retrieved from the NCBI Database, and the sequence alignments are based on one virus strain each (PEDV-YN144: KT021232.1; HCoV-229E: KU291448.1; HCoV-NL63: NC_005831.2; FIPV-WSU-79/1146: AAY32595.1; TGEV-PUR-MAD: NC_038861.1; SARS-CoV-Tor2: NP_828863.1; SARS-CoV-2-Wuhan-Hu-1: YP_009742612.1; MERS-CoV isolate HCoV-EMC/2012: NC_019843.3; IBV-Beaudette: NP_740626.1; PDCoV/USA/Nebraska145/2015: ANI85845.1; SADS-CoV: QJF53984.1). The key residues for 3CLpro activity sites and the compounds' potential binding sites on PEDV are marked with blue arrows at the bottom. The sequences were aligned using ClustalW2, and images were constructed with ESPript3.0.