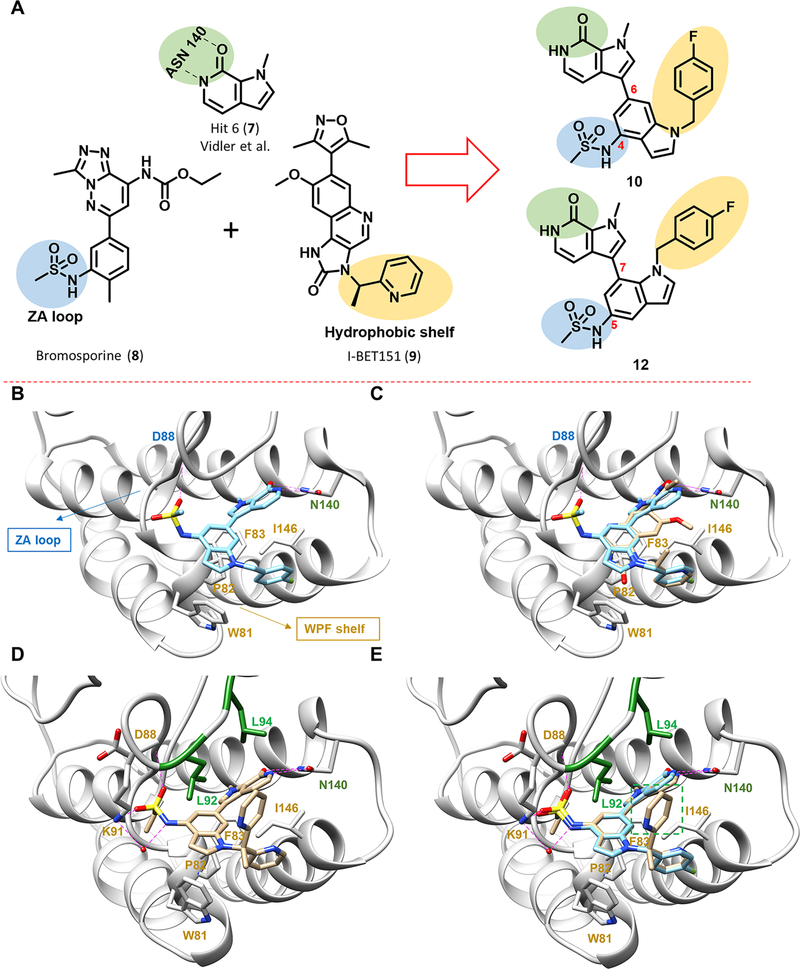
Figure 2.
Design of pyrrolopyridone-based BET inhibitors. (A) Design concept inspired by earlier reported BET inhibitors. Pyrrolopyridone, 7, identified from a virtual screen forms bidentate hydrogen-bonding interaction with Asn140, highlighted in green. The sulfonamide group of 8 interacts with the ZA loop via a putative hydrogen-bonding interaction, highlighted in blue. The heteroaryl moiety of 9 interacts with the hydrophobic WPF shelf, highlighted in yellow. Combining these design parameters leads to the design of 10 and 12 as novel BET inhibitors. (B) Predicted binding pose of compound 10 binding to BRD4-BD1 using Glide (PDB code 3ZYU). Key interacting residues from docking are highlighted and labeled. (C) Overlay of the predicted binding pose of 10 (blue) with 9 (beige) from PDB code 3ZYU. (D) New cocrystal of compound 27 bound to BRD4-BD1 (PDB code 6P05), showing four hydrogen bonds with N140, D88, and K91 and one additional water-mediated hydrogen bond with K91. The “leucine clamp” (L92 and L94) is highlighted in green, and other key interacting residues are highlighted and labeled in red. (E) Overlay of the predicted binding pose of 10 (blue) with the experimentally confirmed binding pose of 27 (beige) clearly showing the second pyridine ring of 27 (boxed in green) that forms a hydrophobic interaction with L92 and L94.