Abstract
We previously reported that bacterial diversity in sputum samples from never-smoking women in rural China varied by lung cancer status and household air pollution (HAP) exposure type. Here, we expand on our associations between environmental exposures and respiratory tract microbiota with an additional 90 never-smoking women from Xuanwei, China. DNA from sputum samples of cases (n = 45) and controls (n = 45) was extracted using a multistep enzymatic and physical lysis, followed by a standardized clean up. V1–V2 regions of 16S rRNA genes were Polymerase chain reaction (PCR) amplified. Purified amplicons were sequenced by 454 FLX Titanium pyrosequencing and high-quality sequences were evaluated for diversity and taxonomic membership. In our population of never-smokers, increased risk of lung cancer was associated with lower alpha diversity compared to higher alpha diversity (Shannon: ORhigh = 1.00 [reference], ORmedium = 3.84 [1.02–14.48], ORlow = 3.78 [1.03–13.82]; observed species: ORhigh = 1.00 [reference], ORmedium = 2.37 [0.67–8.48], ORlow = 2.01 [0.58–6.97]; Phylogenetic Diversity (PD) whole tree: ORhigh = 1.00 [reference], ORmedium = 3.04 [0.85–10.92], ORlow = 2.53 [0.72–8.96]), as well as a decreased relative abundance of Fusobacteria (ORhigh = 1.00 [reference], ORmedium = 1.24 [0.42–3.66], ORlow = 2.01 [0.63–6.44], ptrend = 0.03). Increasing alpha diversity was associated with smoky coal use compared to clean fuel use among all subjects (observed species, P = 0.001; PD whole tree, P = 0.006; Shannon, P = 0.0002), as well as cases (observed species, P = 0.02; PD whole tree, P = 0.03; Shannon, P = 0.03) and controls (observed species, P = 0.01; PD whole tree, P = 0.05; Shannon, P = 0.002). Increased diversity was also associated with presence of livestock (observed species, P = 0.02; PD whole tree, P = 0.02; Shannon, P = 0.03) in the home for cases. Our study is the first to report that decreased microbial diversity is associated with risk of lung cancer. Larger studies are necessary to elucidate the direct and indirect effects attributed to the disease-specific, HAP-specific, and animal-specific associations. Environ. Mol. Mutagen.
Keywords: lung, pulmonary, cancer, bacteria, coal, animal contact, farm
INTRODUCTION
Contrary to previous convention, the lung may not be a sterile environment. For example, bacterial communities have been detected in both lung tissues and bronchoalveolar lavage samples of smokers and nonsmokers (Erb-Downward et al., 2011; Beck et al., 2012; Dickson et al., 2013a, 2013b). Furthermore, etiologic studies suggest that bacteria in the lung may be associated with the risk of nonmalignant respiratory diseases (Lin et al., 2011; Han et al., 2012; Schwabe and Jobin, 2013), such as chronic obstructive pulmonary disease (COPD; Sze et al., 2012).
Previously, in a small exploratory study (Hosgood 3rd et al., 2014), we sought to explore both the role of the lung microbiota in lung cancer among never-smoking women and how household air pollution (HAP), a known lung carcinogen (IARC, 2010), may influence this association. We recruited never-smoking women from the Xuanwei region of China for our study, as this region has the highest female incidence rate of lung cancer in China, and a majority of women in the region are never-smokers with substantial exposure to HAP, attributed to heating and cooking with coal that is often high in polycyclic aromatic hydrocarbon (PAH) content (Mumford et al., 1987; Chapman et al., 1988; Lan et al., 2002; Lan and He, 2004; Hosgood et al., 2008). Our bacterial community survey using 16S rRNA gene sequencing found that there was a significant difference between cases and controls in bacterial diversity in sputum samples. Specifically, Granulicatella, Abiotrophia, and Streptococcus bacteria were enriched in cases compared to controls. Sputum samples also had higher species-level operational taxonomic units (OTUs) in the flora of cases who burned smoky coal in their household for heating and cooking compared to cases who burned smokeless coal.
To follow-up on our previous finding that never-smoking lung cancer cases have different sputum microbiota compared to controls, we conducted a bacterial community survey on an additional 90 never-smoking Xuanwei women. Further, we sought to extend our previous analyses suggesting that sputum microbiota may be influenced by the type of coal burned in the home, by evaluating the associations between sputum microbiota and HAP attributed to a variety of solid fuel types, as well as additional environmental exposures common to Xuanwei residents, such as close contact with farm animals.
MATERIALS AND METHODS
Similar to our initial effort (Hosgood 3rd et al., 2014), cases and controls were selected from an ongoing case–control study of never-smoking females in Xuanwei and Fuyuan counties, China. Cases were women with newly diagnosed primary lung cancer (ICD-9162) who currently lived in Xuanwei and Fuyuan counties and who were evaluated at one of the six study hospitals in this region. Subjects were enrolled between March 2006 and March 2010. The diagnosis of lung cancer was made by histology or cytology. Cases were restricted to those aged 18–79 years of age at the time of diagnosis. Controls were women who had never been diagnosed with lung cancer and were individually matched to cases by age (±5 years) and hospital. Controls were selected from never-smoking female patients aged 18–79 years old who were evaluated at the same hospitals as the cases and who were current residents of Xuanwei or Fuyuan counties. The controls were required to have admission diagnoses of diseases and conditions that were unrelated to the study’s primary hypotheses regarding household coal exposure and air pollution. Control diseases were drawn from a diverse, large number of categories to ensure that no more than 20% of controls had any one condition. During study enrollment, all subjects were administered a standardized questionnaire by a trained interviewer. The questionnaire ascertained information hypothesized to be related to lung cancer risk, such as lifestyle and household characteristics. Biological samples were also collected at the time of enrollment. Sputum samples were collected noninvasively through participant-induced coughing (i.e., without induction) with the expectorate immediately preserved in Saccamono’s fixative, using a patient-induced coughing method that was developed and used successfully during a previous case–control study conducted in Xuanwei (Lan et al., 2000). Cytological examination of sputum collected via this method, specifically from subjects without lung cancer, found that the sputum was derived from the lower respiratory tract and confirmed the presence of bronchial epithelial cells (Keohavong et al., 2005). Buccal cells and sputum samples were collected from study subjects before surgery or other treatment. This study was reviewed and approved by the NIH’s Institutional Review board. All subjects provided written, informed consent.
For the present project, a subgroup of never-smoking female lung cancer cases (n = 45) and never-smoking female controls (n = 45) was randomly selected. The age range of cases and controls selected for this analysis was 34–72 years. Data collected at the time of enrollment were used to categorize cases and controls based on the environmental exposures found in their current homes. First, subjects were categorized based on their respective longest fuel types used in the home: smoky coal, smokeless coal, and other (clean fuels including electricity and gas). The longest fuel type used in the home was determined by the fuel that was used for the most number of years in the subject’s current home. Among the subjects in this analysis, the longest fuel type used accounted for an average of 94.7% of years of fuel use in the current home. Second, subjects were categorized based on animals living in the home: pigs (yes/no), cows and horses (yes/no), chickens (yes/no), and livestock (yes/no).
All sputum and buccal samples were blinded and sent for total genomic DNA extraction using the protocol described by Zupancic et al. (2012). Briefly, samples were pelleted by centrifugation at 10,000 rpm for 15 min, after which the supernatant was removed. Cell lysis was initiated by the addition of 1 mL of phosphate-buffered saline and a first enzymatic cocktail composed of lysozyme, mutanolysin, and lysostaphin. After a 30-min incubation at 37°C, the samples were further lysed by the addition of proteinase K and 10% sodium dodecyl sulfate (SDS), followed by incubation at 55°C for 45 min. Mechanical disruption was then performed by bead beating using a FP120 FastPrep instrument (MP Biomedicals LLC, Santa Ana, CA) and 0.1 mm silica spheres. The resulting crude lysate was processed using the ZYMO Fecal DNA kit (Zymo Research, Irvine, CA) according to the manufacturer’s recommendations. Negative extraction controls were performed to ensure that the samples were not contaminated by exogenous bacterial DNA during the extraction process. Negative extraction controls (phosphate buffered saline (PBS)) were processed in parallel with each extraction to ensure no contaminating DNA was introduced during the DNA extraction process and PCRs. All samples included in our analyses had negative controls. Further, no-template negative controls were processed for each primer pair.
Universal primers (27F, 338R) were then used for PCR amplification of the V1–V2 hypervariable regions of 16S rRNA genes. The 338R primer included a unique sequence tag to barcode each sample (Zupancic et al., 2012). Using 96 barcoded 338R primers, the V1–V2 regions of 16S rRNA genes were amplified in 96-well microtiter plates using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) and 50 ng of template DNA in a total reaction volume of 25 μL, using the cycling conditions described previously (Zupancic et al., 2012). Negative controls without a template were included for each barcoded primer pair. PCR products were quantified using the Quant-iT PicoGreen dsDNA assay (Thermo Fisher Scientific, Waltham, MA), and equimolar amounts (100 ng) of the PCR amplicons were mixed in a single tube. The purified amplicon mixture was then sequenced by 454 FLX Titanium pyrosequencing (Roche/454 Life Sciences) using 454 Life Sciences primer A, using protocols recommended by the manufacturer as amended by the Genomics Resource Center at the Institute for Genome Sciences.
16S rRNA gene sequences output by the sequencer were demultiplexed using barcodes, trimmed of forward and reverse primer sequences, filtered for length and quality, and corrected for homopolymer errors. Passing sequences were characterized for diversity and taxonomic composition using the QIIME (version 1.9.1; Caporaso et al., 2010) and R packages. Sequences were clustered into OTUs with a 97% identity threshold using closed reference OTU picking method with Greengenes 16S database release 13_8. The average number of reads per sample was 6,992 (median = 6,765). Alpha- and beta-diversity metrics and relative abundance tables were computed in QIIME. Alpha-diversity metrics (observed species, PD whole tree, Shannon index) were calculated based on rarefaction depth from 1,000 to 10,000 (Supporting Information Fig. S1). One potential outlier was found to have high alpha-diversity measures relative to all other samples, and thus excluded from subsequent analyses. Results with and without this outlier were similar (data not shown). Beta-diversity analyses (Bray Curtis dissimilarity index, weighted UniFrac distance, unweighted UniFrac distance) were performed after rarefaction to a depth of 1,000 sequences. P-values for each phenotype associated with alpha diversity were calculated by linear regression in R adjusted for age, HAP fuel type, and education. The association with beta-diversity matrix was evaluated using the Microbiome Regression-Based Kernel Association Test (MiRKAT; Zhao et al., 2015), adjusting for age, HAP fuel type, and education. Sputum samples for each case and control were selected for 16S rRNA gene sequencing. Randomly selected buccal samples from randomly selected cases (n = 3) and controls (n = 7) were used to further evaluate the overlap in sputum–buccal pairs from the same subject, which we previously identified (Hosgood 3rd et al., 2014). Similar to our initial effort (Hosgood 3rd et al., 2014), the observed OTU abundances among these 10 sputum–buccal pairs were not highly correlated, with an average Pearson’s correlation coefficient of the relative abundances equal to 0.39 (confidence interval: 0.28–0.50).
RESULTS
Cases and controls were similar in age, village, education, family history of lung cancer, and type of solid fuel used in the home (Table I). Cases were more likely than controls to have lived with pigs in the current home (P = 0.044). In our population of nonsmokers, increased risk of lung cancer was associated with lower alpha diversity compared to higher alpha diversity (Table II; Shannon: ORhigh = 1.00 [reference], ORmedium = 3.84 [1.02–14.48], ORlow = 3.78 [1.03–13.82]; observed species: ORhigh = 1.00 [reference], ORmedium = 2.37 [0.67–8.48], ORlow = 2.01 [0.58–6.97]; PD whole tree: ORhigh = 1.00 [reference], ORmedium = 3.04 [0.85–10.92], ORlow = 2.53 [0.72–8.96]). The unweighted UniFrac distance was also associated with case–control status (P = 0.011); however, the other two beta-diversity metrics were not associated with case–controls status (P = 0.38 for Bray Curtis; P = 0.89 for Weighted UniFrac). Of note, there was a suggestion that increased risk of lung cancer was associated decreased relative abundance of phylum Fusobacteria (ORhigh = 1.00 [reference], ORmedium = 1.24 [0.42–3.66], ORlow = 2.01 [0.63–6.44], ptrend = 0.03) and class Fusobacteriia (ORhigh = 1.00 [reference], ORmedium = 1.24 [0.42–3.66], ORlow = 2.01 [0.63–6.44], ptrend = 0.03; Supporting Information Table SI).
Table 1:
Characteristics of never smoking female lung cancer cases and never smoking female controls from China who participated in this study.
| Controls (n = 45) | Cases (n = 45) | |||||
|---|---|---|---|---|---|---|
| Variables | N | % | N | % | P* | |
| Age (stratified based on median in controls) | <54 years | 19 | 42.2 | 22 | 48.9 | 0.67 |
| ≥54 years | 26 | 57.8 | 23 | 51.1 | ||
| Current county | Xuanwei | 43 | 95.6 | 45 | 100 | 0.47 |
| Fuyuan | 2 | 4.4 | 0 | 0 | ||
| Current village | Laibin | 23 | 51.1 | 23 | 51.1 | 1.00 |
| Non-Laibin | 22 | 48.9 | 22 | 48.9 | ||
| Education | No School | 16 | 35.6 | 21 | 46.7 | 0.34 |
| Elementary school | 18 | 40.0 | 18 | 40.0 | ||
| Middle school and higher | 11 | 24.4 | 6 | 13.3 | ||
| Family history of lung cancer | No | 36 | 80.0 | 32 | 72.7 | 0.58 |
| Yes | 9 | 20.0 | 12 | 27.3 | ||
| Longest fuel used at current home | Smoky coal | 28 | 62.2 | 30 | 66.7 | 0.31 |
| Smokeless coal | 2 | 4.4 | 5 | 11.1 | ||
| Others (clean fuel) | 15 | 33.3 | 10 | 22.2 | ||
| Lived with pigs in current home | No | 20 | 44.4 | 10 | 22.2 | 0.044 |
| Yes | 25 | 55.6 | 35 | 77.8 | ||
| Lived with cows/horses in current home | No | 28 | 62.2 | 21 | 46.7 | 0.20 |
| Yes | 17 | 37.8 | 24 | 53.3 | ||
| Lived with chickens in current home | No | 19 | 42.2 | 11 | 24.4 | 0.12 |
| Yes | 26 | 57.8 | 34 | 75.6 | ||
P-values were obtained using Pearson’s chi-squared test.
Table 2:
Alpha Diversity associated with risk of lung cancer in Xuanwei, China.
| Medium vs. high diversity | Low vs. high diversity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Odds | 95% CI | Odds | 95% CI | ||||||
| Ratio | Lower | Upper | P | Ratio | Lower | Upper | P | p-Trend | |
| Observed species | 2.38 | 0.67 | 8.48 | 0.18 | 2.01 | 0.58 | 6.97 | 0.27 | 0.19 |
| PD whole tree | 3.05 | 0.85 | 10.92 | 0.09 | 2.53 | 0.72 | 8.96 | 0.15 | 0.07 |
| Shannon | 3.84 | 1.02 | 14.48 | 0.05 | 3.78 | 1.03 | 13.82 | 0.04 | 0.22 |
Adjusted for age, coal use and education
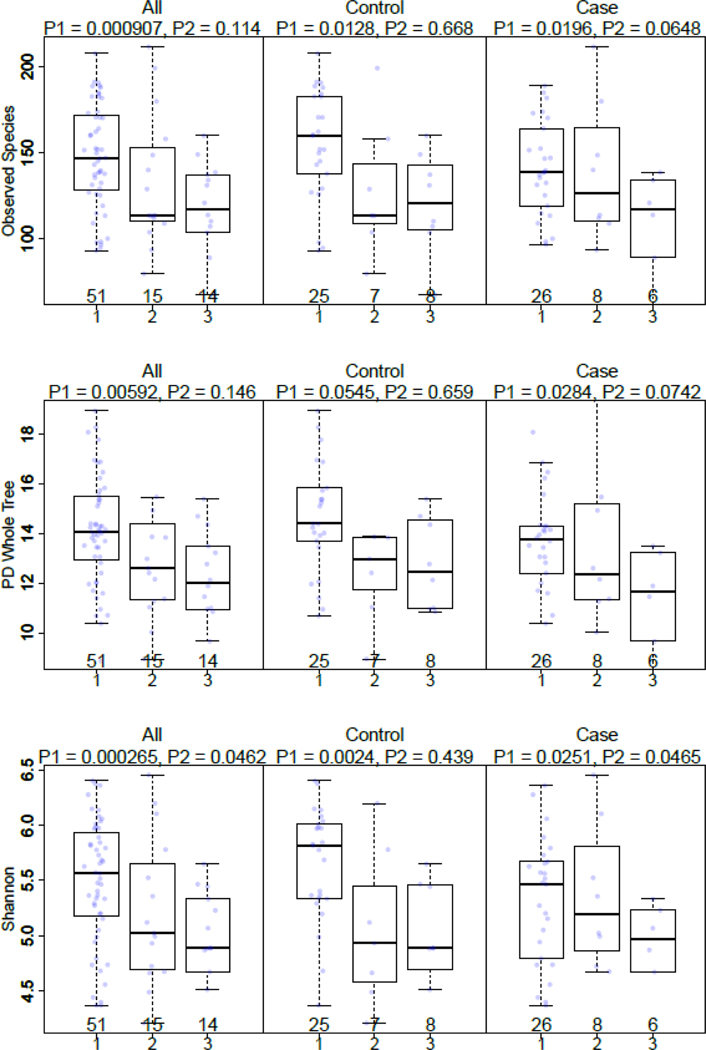
To help understand the relationship between microbiota and the environmental exposures, we evaluated the associations of diversity measures and exposures that were present at the time of sputum collection. When comparing alpha diversity by fuel type used, an increasing microbiome alpha diversity in sputum was associated with smoky coal use compared to clean fuels among all subjects (observed species, P = 0.001; PD whole tree, P = 0.006; Shannon, P = 0.0002), as well as cases (observed species, P = 0.02; PD whole tree, P = 0.03; Shannon, P = 0.03) and controls (observed species, P = 0.01; PD whole tree, P = 0.05; Shannon, P = 0.002; Fig. 1; Supporting Information Table SII). No differences in alpha diversity (P > 0.05) were observed when comparing smokeless coal users and clean fuel users (Fig. 1; Supporting Information Table SII). For all alpha-diversity metrics, fuel type was significantly associated with variance, while age was not (Shannon: fuel type, P = 0.001, age, P = 0.83; observed species: fuel type, P = 0.004, age, P = 0.64; PD whole tree: fuel type, P = 0.025, age, P = 0.30). To further explore the associations with coal type, we evaluated alpha diversity in relation to use of Laibin coal, which has been associated with the highest lung cancer rates (Supplemental Fig. S2). Interestingly, alpha diversity was increased in controls using Laibin coal (observed species, P = 0.003; PD whole tree, P = 0.15; Shannon, P = 0.03).
Figure 1: Alpha diversity by type of fuel type used in the home*, by case status.
*Smoky coal (1), smokeless coal (2), and clean fuels (3); P1: smoky coal (1) vs clean fuels (3), adjusted for age and education; P2: smokeless coal (2) vs clean fuels (3), adjusted for age and education
In addition to the unique HAP-attributed exposures, some residents of Xuanwei live with farm animals in the home. Microbiome alpha diversity (measured by observed species) in sputum was associated with the presence of any livestock in the household among cases (observed species, P = 0.02; PD whole tree, P = 0.02; Shannon, P = 0.03), and potentially the presence of cows or horses and pigs (Supporting Information Table SII). Beta diversity was not associated with presence of animals (Supporting Information Table SIII). Given the robustness of our metadata, we were able to conduct exploratory analyses of the associations between each phylum and class with environmental exposures (Supporting Information Table SIV). These results suggest that increased abundance of class Fusobacteriia (ptrend = 0.03 in all subjects; ptrend = 0.05 in cases) and decreased abundance of class Betaproteobacteria (ptrend = 0.01 in cases) are associated with smoky coal use compared to using clean fuels. Decreased abundance of class Betaproteobacteria (ptrend = 0.02 in all subjects; ptrend = 0.02 in cases) was also associated with smokeless coal use compared to using clean fuels. Decreased abundance of phylum Acidobacteria (ptrend = 0.02 in all subjects; ptrend = 0.05 in controls) and class Chloracidobacteria (ptrend = 0.01 in all subjects; ptrend = 0.05 in controls) was associated with the presence of livestock.
DISCUSSION
We conducted a case–control study of lung cancer among nonsmoking females in rural China and found that decreased diversity in the lung microbiome was associated with increased risk of lung cancer. Our observed suggestion of microbiome diversity differences in sputum among lung cancer cases compared to controls is similar to that observed in our previous small exploratory study (Hosgood 3rd et al., 2014). This study adds to the emerging hypothesis that the respiratory tract microbiota may contribute to the risk of respiratory diseases, such as asthma and COPD (Sze et al., 2012; Burbank et al., 2017; Jackson et al., 2017). While no other study has yet to report that decreased diversity is associated with increased risk of lung cancer, other research groups have observed (1) that malignant lung cancer tissues have lower alpha diversity compared to nonmalignant tissues (Yu et al., 2016), and (2) decreased alpha diversity in sputum has been shown to be associated with increased severity of certain types of COPD exacerbations (Wang et al., 2018).
The respiratory tract microbiota is the first point of contact with inhalable exposures, and may influence the body’s ability to process and respond to environmental exposures (Backhed, 2012). Microbiotas have been shown to detoxify PAHs (Backhed, 2012; Kostic et al., 2013), which are a class of lung carcinogens in HAP (IARC, 1983; Mumford et al., 1993). A major strength of the current study was our ability to conduct detailed analyses of the relationship between microbiome diversity and HAP. Notably, diversity increased depending on HAP fuel type. Further, we observed bacterial composition differences associated with specific coal subtypes. Our analysis of Laibin coal is of interest as exposure to smoke attributed to Laibin coal has been associated with some of the highest risks of lung cancer in the world, often overwhelming the effect of tobacco smoke on risk of lung cancer (Lan et al., 2002; Hosgood et al., 2008). These HAP– microbiome associations are strengthened when observed in analyses restricted to controls, as they are void of reverse causality due to lung cancer.
In rats, exposure to biomass smoke has been shown to increase bacterial abundance and diversity in lung tissue, and particulate matter exposure significantly increased the capacity of alveolar macrophages to phagocytose bacteria (Li et al., 2017). Recently, a cross-sectional study of bronchoalveolar lavage samples from 44 healthy adults in Malawi evaluated that association between HAP and the lung microbiome (Rylance et al., 2016). Similar to the current study, microbial differences were observed between subjects burning solid fuel compared to those using nonsolid fuels (e.g., electricity). Subjects with high HAP-attributed particulate exposure had significantly higher abundance of Neisseria and Streptococcus compared to subjects with low exposure. Interestingly, our original study observed higher abundance of Neisseria and Streptococcus in cases compared to controls (Hosgood 3rd et al., 2014); however, the current study did not replicate this finding. Together these studies suggest that exposure to solid fuel combustion byproducts is associated with increased bacterial diversity, potentially enriched for Streptococcus, which has been causally linked to pneumonia (van der Poll and Opal, 2009). The current study provides additional mechanistic clues into the potential bacteria associated with nonsmoking lung cancer. We found that among nonsmokers, subjects with decreased relative abundance of phylum Fusobacteria experienced an increased risk of lung cancer. Increased abundance of class Fusobacteriia was associated coal type used in the homes of our subjects. Although we are the first to report these associations with Fusobacteria, to the best of our knowledge, it is consistent with recent studies that have found lung-derived Fusobacteria to be associated with sleep apnea (Lu et al., 2018) and asthma (Millares et al., 2017).
Our study has a number of strengths and limitations that warrant consideration. First, our population is a major strength. We recruited females from the unique population of Xuanwei, who have substantial HAP exposures, and used only never-smokers to minimize confounding from active tobacco smoking. Second, we used sophisticated exposure assessments to ascertain the current HAP exposures at the time of biological sample collection. A limitation of our study is that the 16S RNA gene-based sequencing assay does not differentiate between viable and nonviable bacteria. As such, we are not able to determine if our observed perturbations are attributed to living, metabolically active bacterial cells or inactivated bacteria that are in the process of being eliminated by the lung’s clearance mechanism. Finally, given our limited sample size, our results should be interpreted with caution, particularly the associations between individual taxa and specific environmental exposures (Supporting Information Table SIV).
Although our findings must be replicated in larger sample sizes, they highlight the need for further research related to the role of bacterial communities in respiratory health and respiratory diseases, potentially leading to a novel field of research related to the role that pathogens play in the lung.
Future studies replicating our findings should focus on prospectively collected serial samples, and potentially consider evaluating the microbiota of the gut, which has been suggested to be associated with immune system’s modulation of the microbiome composition at mucosal surfaces, epithelial barrier function, and local immunity response in the lung (Smits et al., 2016). Interestingly, an analysis of ~1,000 healthy individuals found significant similarities in the compositions of the gut microbiomes of genetically unrelated individuals who share a household, and that over 20% of the interperson microbiome variability is associated with factors related to environmental and anthropometric factors (Rothschild et al., 2018). Further elucidating the relationship between HAP and microbiota may provide etiologic clues to the underlying mechanisms of HAP-attributed diseases, which is of immense public health importance given that HAP is the seventh leading cause of disease in the world (Global Burden of Disease Collaborators, 2017). This field of research could potentially identify subpopulations of HAP-attributed diseases and lead to novel public health interventions (Adar et al., 2016).
Supplementary Material
ACKNOWLEDGMENT
The Intramural National Cancer Institute (N01-CO12400) program funded the work.
Grant sponsor: National Cancer Institute; Grant number: N01-CO-12400.
Footnotes
Additional Supporting Information may be found in the online version of this article.
CONFLICTOFINTEREST
None to declare.
REFERENCES
- Adar SD, Huffnagle GB, Curtis JL. 2016. The respiratory microbiome: An underappreciated player in the human response to inhaled pollutants? Ann Epidemiol 26:355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F. 2012. Host responses to the human microbiome. Nutr Rev 70 (Suppl 1):S14–S17. [DOI] [PubMed] [Google Scholar]
- Beck JM, Young VB, Huffnagle GB. 2012. The microbiome of the lung. Transl Res 160:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank AJ, Sood AK, Kesic MJ, Peden DB, Hernandez ML. 2017. Environmental determinants of allergy and asthma in early life. J Allergy Clin Immunol 140:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RS, Mumford JL, Harris DB, He ZZ, Jiang WZ, Yang RD. 1988. The epidemiology of lung cancer in Xuan Wei, China: Current progress, issues, and research strategies. Arch Environ Health 43:180–185. [DOI] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Huffnagle GB. 2013a. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med 7: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Huang YJ, Martinez FJ, Huffnagle GB. 2013b. The lung microbiome and viral-induced exacerbations of chronic obstructive pulmonary disease: New observations, novel approaches. Am J Respir Crit Care Med 188:1185–1186. [DOI] [PubMed] [Google Scholar]
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. 2011. Analysis of the lung microbiome in the “healthy” smoker and in copd. PLoS One 6:e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Collaborators. 2017. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet 390:1345–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, et al. 2012. Significance of the microbiome in obstructive lung disease. Thorax 67: 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, Chapman R, Shen M, Blair A, Chen E, Zheng T, Lee KM, He X, Lan Q. 2008. Portable stove use is associated with lower lung cancer mortality risk in lifetime smoky coal users. Brit J Cancer 99:1934–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD 3rd, Sapkota AR, Rothman N, Rohan T, Hu W, Xu J, Vermeulen R, He X, White JR, Wu G, et al. 2014. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen 55:643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. 1983. Polynuclear Aromatic Compounds. Lyon: International Agency for Research on Cancer Monographs on the Evaluation of the Carcinogenic Risks to Humans. [Google Scholar]
- IARC. 2010. Household Use of Solid Fuels and High-Temperature Frying. Lyon: World Health Organization. [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, Gern JE, Lemanske RF Jr. 2017. Lessons learned from birth cohort studies conducted in diverse environments. J Allergy Clin Immunol 139:379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P, Lan Q, Gao WM, Zheng KC, Mady HH, Melhem MF, Mumford JL. 2005. Detection of p53 and k-ras mutations in sputum of individuals exposed to smoky coal emissions in Xuan Wei county, China. Carcinogenesis 26:303–308. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27: 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, He X, Costa DJ, Tian L, Rothman N, Hu G, Mumford JL. 2000. Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes, and lung cancer risk: A case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev 9:605–608. [PubMed] [Google Scholar]
- Lan Q, Chapman RS, Schreinemachers DM, Tian L, He X. 2002. Household stove improvement and risk of lung cancer in Xuan Wei, China. J Natl Cancer Inst 94:826–835. [DOI] [PubMed] [Google Scholar]
- Lan Q, He XZ. 2004. Molecular epidemiological studies on the relationship between indoor coal burning and lung cancer in Xuan Wei, China. Toxicology 198:301–305. [DOI] [PubMed] [Google Scholar]
- Li N, He F, Liao B, Zhou Y, Li B, Ran P. 2017. Exposure to ambient particulate matter alters the microbial composition and induces immune changes in rat lung. Respir Res 18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KW, Li J, Finn PW. 2011. Emerging pathways in asthma: Innate and adaptive interactions. Biochim Biophys Acta 1810:1052–1058. [DOI] [PubMed] [Google Scholar]
- Lu D, Yao X, Abulimiti A, Cai L, Zhou L, Hong J, Li N. 2018. Profiling of lung microbiota in the patients with obstructive sleep apnea. Medicine 97:e11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millares L, Bermudo G, Perez-Brocal V, Domingo C, Garcia-Nunez M, Pomares X, Moya A, Monsó E. 2017. The respiratory microbiome in bronchial mucosa and secretions from severe IgE-mediated asthma patients. BMC Microbiol 17:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford JL, He XZ, Chapman RS, Cao SR, Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC. 1987. Lung cancer and indoor air pollution in Xuan Wei, China. Science 235:217–220. [DOI] [PubMed] [Google Scholar]
- Mumford JL, Lee XM, Lewtas J, Young TL, Santella RM. 1993. DNA adducts as biomarkers for assessing exposure to polycyclic aromatic-hydrocarbons in tissues from Xuan Wei women with high exposure to coal combustion emissions and high lung-cancer mortality. Environ Health Perspect 99:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. [DOI] [PubMed] [Google Scholar]
- Rylance J, Kankwatira A, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, Sodergren E, Weinstock GM, et al. 2016. Household air pollution and the lung microbiome of healthy adults in Malawi: A cross-sectional study. BMC Microbiol 16:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Jobin C. 2013. The microbiome and cancer. Nat Rev Cancer 13:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits HH, van der Vlugt LE, von Mutius E, Hiemstra PS. 2016. Childhood allergies and asthma: New insights on environmental exposures and local immunity at the lung barrier. Curr Opin Immunol 42:41–47. [DOI] [PubMed] [Google Scholar]
- Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. 2012. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 185:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Poll T, Opal SM. 2009. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374:1543–1556. [DOI] [PubMed] [Google Scholar]
- Wang Z, Singh R, Miller BE, Tal-Singer R, Van Horn S, Tomsho L, Mackay A, Allinson JP, Webb AJ, Brookes AJ, et al. 2018. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 73:331–338. [DOI] [PubMed] [Google Scholar]
- Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, Caporaso NE, Goedert JJ, Ravel J, Landi MT. 2016. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou JJ, Ringel Y, Li H, Wu MC. 2015. Testing in microbiome-profiling studies with mirkat, the microbiome regression-based kernel association test. Am J Hum Genet 96:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, Jones C, Knight R, Walters WA, Knights D, et al. 2012. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One 7:e43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.