Figure 7.
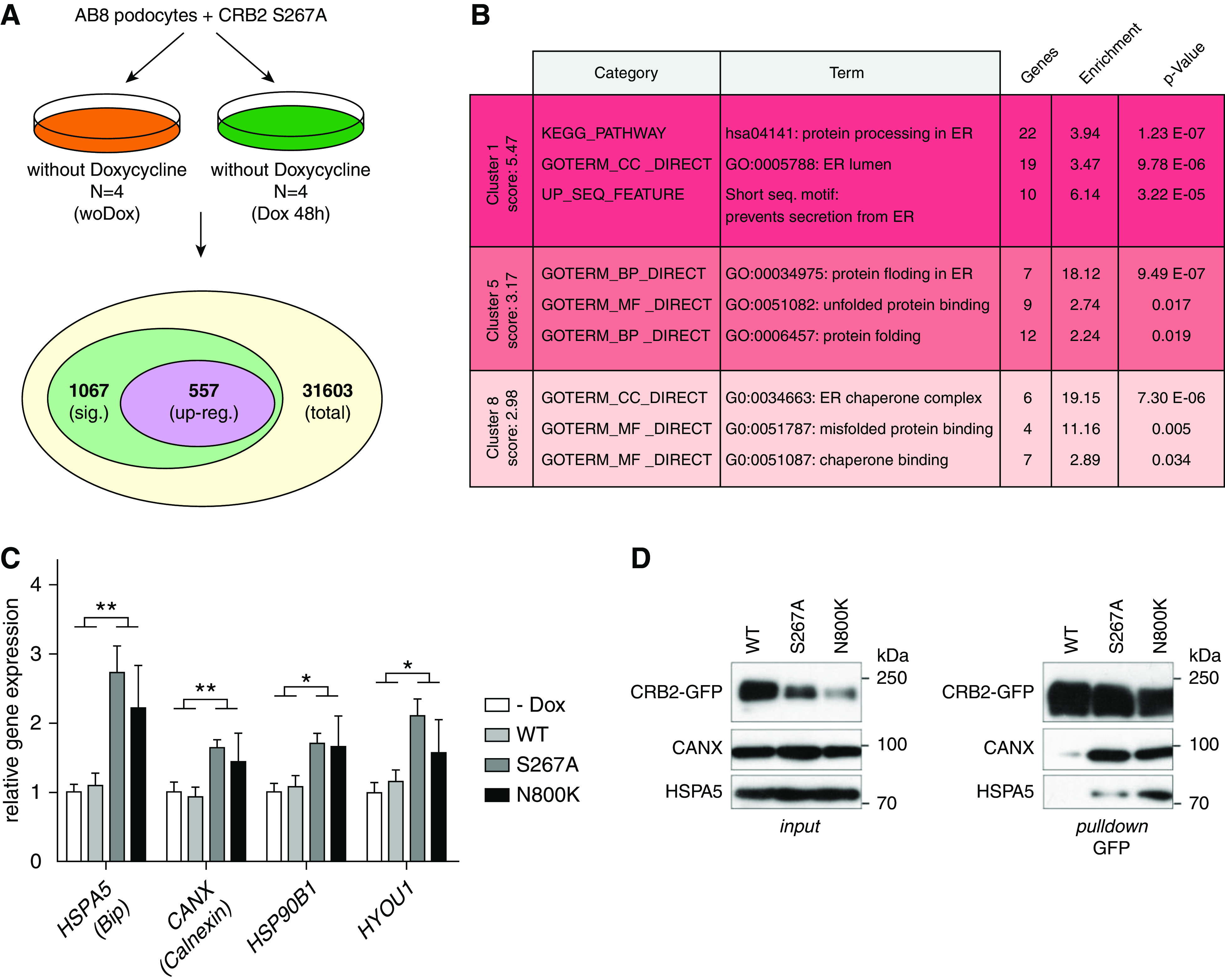
ER-localized CRB2 mutants cause ER stress. (A) RNAseq analyses of AB8 podocyte cells expressing the GFP-tagged CRB2 S267A mutant (with Dox) compared with noninduced control (woDox) resulted in 1067 differentially expressed genes (sig.). (B) Functional gene enrichment analyses (DAVID bioinformatics) of upregulated genes (upreg.; 557) revealed activation of ER stress and ER stress–associated pathways. (C) Quantitative real-time PCR analyses of ER stress genes HSPA5 (BiP, Grp78), CANX (Calnexin), HSP90B1 (Grp94) and HYOU1 (Grp170, Orp150): ER stress was only induced in cells expressing the S267A or N800K mutant. Noninduced cells (−Dox) or cells expressing wild-type CRB2 (WT) did not show upregulation of these genes. Unpaired two-tailed t test. n≥5. *P=0.05; **P=0.01. (D) GFP pull-down assays: CRB2 S267A and N800K mutants bind stronger to ER-localized protein folding chaperones HSPA5 (BiP) and CANX (Calnexin) than WT. (n=3).
