Abstract
Two new species of Fulvifomes are described from specimens collected in rainforests of Nonggang Nature Reserve of southern China, based on morphological characteristics and molecular phylogenetic analysis of the internal transcribed spacer (ITS) and nuclear large subunit ribosomal DNA (nLSU) sequences. Fulvifomes nonggangensis sp. nov. is characterized by perennial, sessile and solitary basidiocarps, applanate pileus, small cystidioles of 9.9–15.4 × 2.9–3.5 μm, large pores of 5–6 per mm, a dimitic hyphal system, and broadly ellipsoid basidiospores of 4.3–5.3 × 3.3–4.2 μm. F. tubogeneratus sp. nov. is characterized by perennial, sessile, and imbricate basidiocarps, a duplex context, small pores of 7–8 per mm, a dimitic hyphal system, and ovoid to subglobose basidiospores of 5.72 × 5.00 μm.
Keywords: Hymenochaetaceae, morphology, phylogenetic, Taxonomy
1. Introduction
Most species in the family Hymenochaetaceae were of medicinal value, while some were plant pathogens causing a white rot [1–3]. Fulvifomes Murrill was established by Murrill in 1914 and typified with F. robiniae Murrill to accommodate species with perennial and sessile basidiocarps, sulcate surface, ungulate or applanate pilei and smooth, ferruginous or fulvous spores in the family Hymenochaetaceae [4]. Fulvifomes comprises 28 species according to Index Fungorum (http://www.indexfungorum.org/Names/Names.asp, accessed on 2020/10/27). Fulvifomes was considered as a synonym of Phellinus Quél. for several decades by Ryvarden and other mycologists [5–8], until Wagner and Fischer [9] provided evidence to confirm Fulvifomes as an independent generic rank within Hymenochaetaceae based on molecular phylogenetic analyses. Furthermore, the genus Aurificaria, represented by Aurificaria luteoumbrina, was very close to Phylloporia and Fulvifomes reflected by the phylogenetic trees. Zhou [10] re-delimited the circumscription of Fulvifomes based on phylogenies inferred from nuclear large subunit ribosomal DNA (nLSU) and internal transcribed spacer (ITS) regions. Hattori et al. [11] also provided the key to worldwide species of Fulvifomes. Recently, still several new species were included within Fulvifomes [12–15].
During the macrofungal diversity survey in southern China, two additional undescribed species of Fulvifomes were found and identified as new by morphological characteristics and phylogenetic analysis inferred from the ITS and nLSU regions.
2. Materials and methods
2.1. Morphological studies
Specimens in this study were deposited in the herbarium of Guangxi University (GXU). The method of microscopic procedure followed Dai [2]. Special color definition followed Ridgway [16]. Sections were studied at magnification up to × 1500 using a Nikon Eclipse 80i microscope (Nikon Corporation, Tokyo, Japan). Abbreviations were used in text: IKI: Melzer’s reagent; IKI−: negative in Melzer’s reagent; KOH: 5% potassium hydroxide; CB: cotton blue; CB+: cyanophilous; CB−: acyanophilous; L: mean spore length (arithmetic average of all spores); W: mean spore width (arithmetic average of all spores); Q: variation in the L/W ratios between the specimens studied; n: number of spores measured from given number of specimens.
2.2. DNA extraction, PCR, and sequencing
DNA extraction followed the protocol of conventional cetyl trimethylammonium bromide (CTAB) method. Nuclear ITS and nLSU regions were amplified with primer pairs ITS5/ITS4 [17] and LR0R/LR5 [18]. The PCR products were directly purified and sequenced by Beijing Genomics Institute (BGI; Shenzhen, China). PCR procedure was followed as: initial denaturation at 94 °C for 5 min, followed by 30 cycles at 94 °C for 40 s, 56 °C for 40 s, and 72 °C for 1 min, and a final extension of 72 °C for 10 min.
2.3. Phylogenetic analysis
In this study, eight new sequences were generated and additional sequences were obtained from GenBank are listed in Table 1. The ITS datasets with Fomitiporella inermis as outgroup, while the ITS + nLSU datasets with Oninia tomentosa, respectively.
Table 1.
Information for sequences used in this study.
| Species | Geographic origin | Strain no. | GenBank accessions |
|
|---|---|---|---|---|
| ITS | nLSU | |||
| Fulvifomes elaeodendri | South Africa | CMW47808 | MH599093 | MH599131 |
| Fulvifomes elaeodendri | South Africa | CMW47909 | MH599096 | MH599132 |
| Fulvifomes fastuosus | Thailand | LWZ 20140801-1 | KR905675 | – |
| Fulvifomes fastuosus | Viet Nam | Dai 18292 | MH390411 | – |
| Fulvifomes fastuosus | Philippines | CBS 213.36 | AY558615 | AY059057 |
| Fulvifomes fastuosus | Viet Nam | Dai 18292 | MH390411 | MH390381 |
| Fulvifomes grenadensis | USA | JV1212 2 J | KX960756 | – |
| Fulvifomes grenadensis | Costa Rica | 1607 66 | KX960758 | – |
| Fulvifomes grenadensis | Brazil | JRF74 | MH048097 | MH048087 |
| Fulvifomes grenadensis | Brazil | PH6 | MH048096 | MH048086 |
| Fulvifomes hainanensis | China | Dai 11573 | KC879263 | JX866779 |
| Fulvifomes imbricatus | Thailand | IFP LWZ 20140728-16 | NR_154003 | NG_068762 |
| Fulvifomes imbricatus | Thailand | LWZ 20140729-26 | KR905679 | KR905671 |
| Fulvifomes imbricatus | Thailand | MRNo309 | LC176748 | – |
| Fulvifomes indicus | Zimbabwe | O 25034 | KC879262 | – |
| Fulvifomes kawakamii | Brazil | PPT152 | MH048095 | – |
| Fulvifomes krugiodendri | USA | JV1008 21 | KX960761 | KX960767 |
| Fulvifomes krugiodendri | USA | JV0904_1 | KX960762 | KX960765 |
| Fulvifomes nilgheriensis | Brazil | 3028 | MH390431 | – |
| Fulvifomes nonggangensis | China | GXU1127 | MT571504 | MT571502 |
| Fulvifomes nonggangensis | China | GXU2254 | MT571503 | MT571501 |
| Fulvifomes rhytiphloeus | Brazil | AMO763 | MH048091 | MH048081 |
| Fulvifomes siamensis | Thailand | STRXG2 | JX104708 | JX104755 |
| Fulvifomes siamensis | Thailand | KBXG3 | JX104706 | JX104753 |
| Fulvifomes siamensis | Viet Nam | Dai 18309 | MH390434 | MH390389 |
| Fulvifomes XHJ-2018b | China | Dai 17203 | MH390419 | MH390397 |
| Fulvifomes XHJ-2018b | China | Dai 17470 | MH390418 | MH390395 |
| Fulvifomes XHJ-2018e | USA | JV 0904/65 | MH390422 | – |
| Fulvifomes XHJ-2018e | USA | JV 0312/23.1 | MH390423 | – |
| Fulvifomes XHJ-2018e | USA | JV 0904/76 | MH390424 | – |
| Fulvifomes XHJ-2018h | China | Dai 9642 | MH390429 | MH390379 |
| Fulvifomes XHJ-2018h | China | Dai 10809 | MH390428 | MH390378 |
| Fulvifomes XHJ-2018i | USA | JV 0904/68 | MH390408 | – |
| Fulvifomes XHJ-2018i | USA | JV 1109/77 | MH390409 | – |
| Fulvifomes XHJ-2018l | China | Dai 17911 | MH390405 | – |
| Fulvifomes XHJ-2018l | China | Dai 17917 | MH390406 | – |
| Fulvifomes squamosus | Peru | CS385 | MF479268 | MF479265 |
| Fulvifomes squamosus | Peru | CS456 | MF479267 | MF479266 |
| Fulvifomes thailandicus | Thailand | IFP LWZ 20140731-1 | NR154002 | – |
| Fulvifomes thailandicus | Thailand | LWZ 20140731-1 | KR905672 | KR905665 |
| Fulvifomes tubogeneratus | China | GXU2468 | MT580805 | MT580800 |
| Fulvifomes tubogeneratus | China | GXU2478 | MT580806 | MT580801 |
| Fulvifomes yoroui | Benin | OAB0097 | MN017126 | MN017120 |
| Fomitiporella inermis | USA | JV 1109/19 A | KX181304 | – |
| Fomitiporella inermis | USA | JV 1009/56 | KX181306. | KX181347 |
| Fomitiporella inermis | USA | JV 0509/57 K | KX181305 | KX181346 |
| Fomitiporia tsugina | USA | TOL2-1 | KC551821 | KC551843 |
| Fomitiporia tsugina | USA | TOL2-3 | KC551823 | KC551845 |
| Inonotus porrectus | CAW-30 | HQ589219 | – | |
| Inonotus porrectus | CAW-31 | HQ589220 | – | |
| Inonotus rigidus | China | Cui 8588 | KX674579 | – |
| Inonotus rigidus | China | Cui 8465 | KX674580 | – |
| Onnia tomentosa | Canada | Bud-551-C-1 | JX110072 | JX110116 |
| Phellinus merrillii | PM950703-1 clone 2 | EU035311 | – | |
| Phellinus merrillii | PM950703-1 clone 3 | EU035312 | – | |
| Phellinus robiniae | USA | CBS 211.36 | AY558646 | AY059038 |
| Phellinus robiniae | USA | CFMR:2693 | KX065961 | KX065995 |
| Phellinus robiniae | USA | CFMR:2735 | KX065962 | KX065996 |
| Phellinus piptadeniae | Brazil | MF008 | KP412289 | – |
| Phellinus piptadeniae | Brazil | MF027 | KP412291 | – |
| Phellinus piptadeniae | Brazil | MF034 | KP412295 | KP412276 |
| Phellinus piptadeniae | Brazil | MF036 | KP412297 | KP412277 |
| Phellinus piptadeniae | Brazil | MF038 | KP412299 | KP412278 |
| Phylloporia ephedrae | Turkmenistan | 13690 | MH151184 | – |
| Phylloporia gutta | China | Cui6945 | MH151182 | – |
| Phylloporia gutta | China | Dai16070 | MH151183 | – |
| Tropicoporus boehmeriae | Thailand | LWZ 20140729-10 | KT223640 | – |
| Tropicoporus boehmeriae | Thailand | LWZ 20140729-13 | KT223641 | – |
Sequence datasets of the ITS and the combinability of ITS + nLSU were aligned with MEGA X version 10.0.5 (Pennsylvania State University, PA, USA) [19] and Clustalx version 1.83 (Information Retrieval, London, England) [20], respectively. Sequence alignment was deposited at TreeBASE (Study Accession URL: http://purl.org/phylo/treebase/phylows/study/TB2:S26455) and was executed by paupwin32_4b4a (Sinauer Associates, Sunderland, MA, USA) [21], MrMtgui version 1.0 (http://www.genedrift.org/mtgui.php) and MrModeltest version 2.3 (Uppsala University, Uppsala, Sweden) [22,23] to find the best-fit model for further analysis: MrBayesian analyses were performed by MrBayes version 3.2.2 (University of Rochester, NY, USA) [24] with 5,000,000 generations. Phylogenetic tree of maximum parsimony analyses which performed in PAUP* version 4.0b4a (Sinauer Associates, Sunderland, MA, USA) was generated using tree-bisection reconnection (TBR) branch-swapping algorithm, clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates. Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated. Phylogenetic trees were edited by TreeGraph version 2.3.0-425 beta (BioMed Central, London, England) [25].
Algorithms of two phylogenetic analyses generated nearly congruent topologies for each dataset, only the topology from the MP analysis was presented along with statistical values from the MP and BI algorithms (BS not less than 50% and BPP not less than 0.5) at the nodes.
3. Results
3.1. Phylogenetic analysis
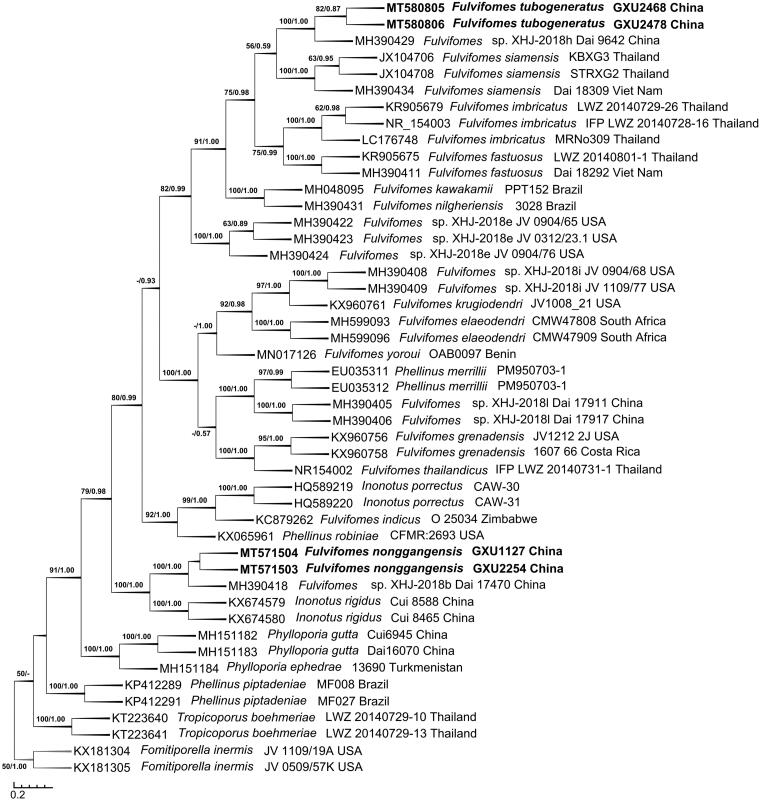
The ITS dataset included sequences from 47 fungal specimens representing 27 taxa. The consensus tree is shown in Figure 1 (TL = 1200, CI = 0.6550, RI = 0.8488, RC = 0.5560, and HI = 0.3450). Best model for the ITS dataset estimated and applied in the Bayesian analysis: HKY + I + G, Lset nst = 2 rates = invgamma, Prset statefreqpr = dirichlet (1,1,1,1). The average standard deviation of split frequencies of Bayesian analysis is 0.004929. Tree topology of the maximum parsimony analysis showed almost same as the tree from Bayesian analysis. The phylogeny based on the ITS dataset (Figure 1) showed that the new species Fulvifomes tubogeneratus clustered with a sequence of F. XHJ-2018h Dai 9642 up to a higher support (BS = 100, BPP = 1.00), then they clustered with F. siamensis to form a small group which evidently fell into the core of Fulvifomes clade; new species F. nonggangensis clustered with F. XHJ-2018b Dai 17470 up to a higher support (BS = 100, BPP = 1.00), then they also clustered with Inonotus rigidus up to a higher support (BS = 100, BPP = 1.00) to form another small group but did not fall into the core of Fulvifomes clade.
Figure 1.
Phylogenetic tree was generated using maximum parsimony analyses based on ITS sequences. Bootstrap values (before the/) higher than 50% and Bayesian posterior probabilities (after the/) more than 0.50 are indicated along the branches.
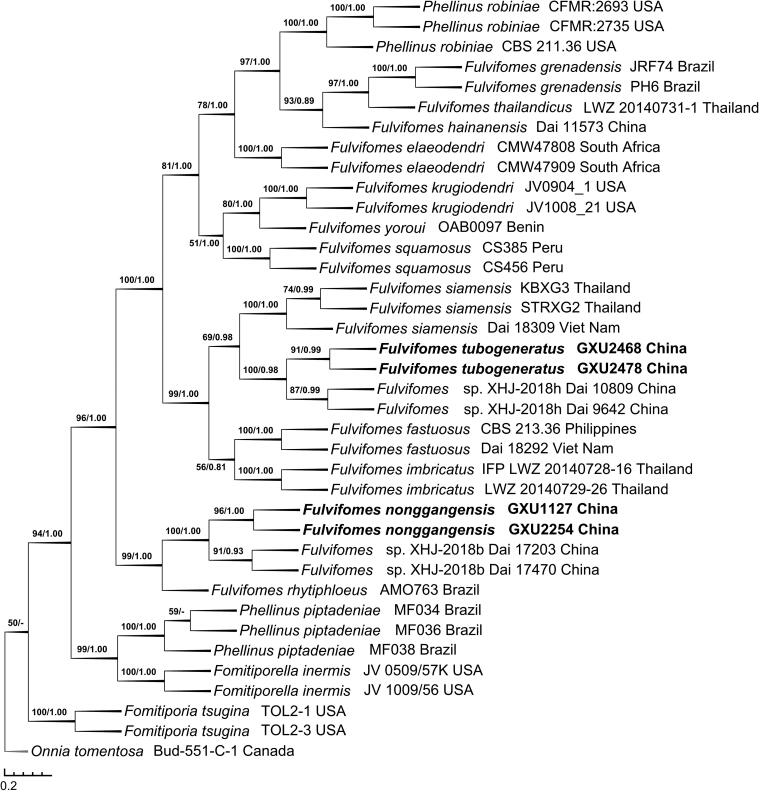
The ITS + nLSU dataset included sequences from 38 fungal specimens representing 20 taxa. The consensus tree is shown in Figure 2 (TL = 1613, CI = 0.6820, RI = 0.8400, RC = 0.5729, and HI = 0.3180). Best model for the ITS + nLSU dataset estimated and applied in the Bayesian analysis: GTR + I + G, Lset nst = 6 rates = invgamma, Prset statefreqpr = dirichlet (1,1,1,1). The average standard deviation of split frequencies of Bayesian analysis is 0.003023. Tree topology of the maximum parsimony analysis showed almost same as the tree from Bayesian analysis.
Figure 2.
Phylogenetic tree was generated using maximum parsimony analyses based on combined ITS + nLSU sequences. Bootstrap values (before the/) higher than 50% and Bayesian posterior probabilities (after the/) more than 0.50 are indicated along the branches.
In the phylogeny inferred from the ITS + nLSU dataset (Figure 2), F. tubogeneratus fell into the core of Fulvifomes clade, similarly to the phylogeny inferred from the ITS dataset, F. tubogeneratus clustered with F. XHJ-2018h Dai 9642 and Dai 10809 up to a higher support (BS = 100, BPP = 0.98), and then clustered with F. siamensis to form a small group; meanwhile, F. nonggangensis clustered with F. XHJ-2018b Dai 17203 and Dai 17470 with a higher support (BS = 100, BPP = 1.00), then they also grouped with F. rhytiphloeus with high support (BS = 99, BPP = 1.00).
3.2. Taxonomy
Fulvifomes nonggangensis F.C. Huang, H.F. Zheng & Bin Liu, sp. nov. (Figures 3 and 4).
Figure 3.
Basidiocarps of Fulvifomes nonggangensis. A: pileal surface of a mature basidiocarp; B: tube surface of a mature basidiocarp; C: a young basidiocarp; D: two aged basidiocarps. Scales bar: 1 cm.
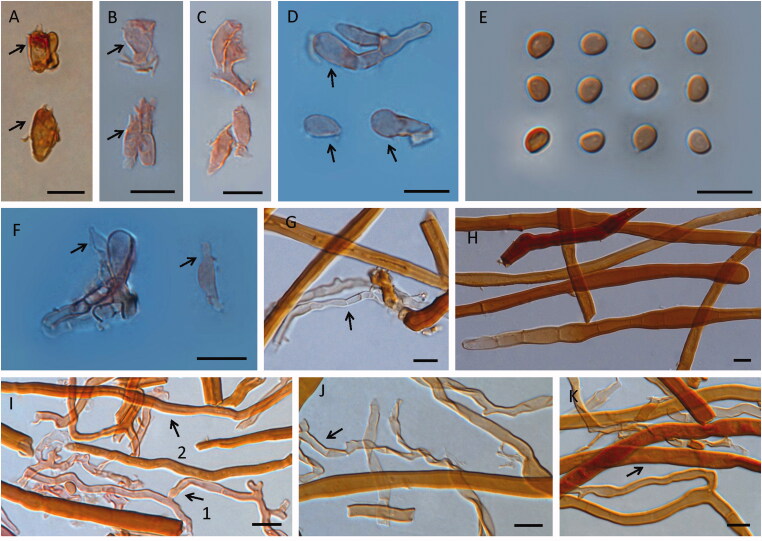
Figure 4.
Microscopic structures of Fulvifomes nonggangensis. Scales bar: 10 μm. A, B, C: basidia; D: basidioles; E: basidiospores; F: cystidioles (arrow pointed); G: generative hyphae from tomentum (arrow pointed); H: skeletal hyphae from tomentum; I: 1, tramal generative hyphae, 2, tramal skeletal hyphae; J: generative hyphae from context (arrow pointed); K: skeletal hyphae from context (arrow pointed).
MycoBank: MB835790
Etymology: nonggangensis (Lat.): referring to the locality of the type specimen.
Type: China, Guangxi Autonomous Region, Chongzuo, Longzhou County, Nonggang Nature Reserve, on living trunks of angiosperm tree, September 19 2012, GXU1127 (Holotype in GXU).
rDNA sequences ex holotype: MT571504 (ITS), MT571502 (nLSU).
Description: Basidiocarps perennial, sessile, solitary, occasionally smaller pileus fused along adjacent margins with other, broadly attached on living trunk, and old fruit body also found on dead trunk, without odor or taste, woody hard. Pileus applanate, of old fruit body (4–5 years) projecting up to 11.1 cm, 14.1 cm wide, and 6.4 cm thick at base. Pileal surface capucine orange, orange to amber brown when fresh, capucine yellow to orange at the actively growing part, orange to antique brown, argus brown when dry, densely tomentose, up to 0.1 cm thick, rough, nodulose, separated by a dense black line tissue from context, concentrically sulcate indistinct to distinct, part of tomentum becoming thinner, nodulose less, and sulcate zone more distinct with age, margin capucine yellow to orange when fresh and capucine orange to raw sienna when dry, obtuse. Pilei of old basidiocarps (4–5 years) argus brown, raw umber to almost black, concentrically sulcate zone distinct, margin narrow, and each year re-expanding from the position beneath the margin forming by previous year, making peripheral part usually looked like slowly descending ladders, and the periphery also radially cracked. Context up to 2.8 cm thick, apricot yellow to orange citrine, woody hard, occasionally a few black line tissues randomly distributed in context. Context of old basidiocarps (4–5 years) up to 1.5 cm thick, carob brown to chestnut brown, and its tomentum becoming a thin layer. Tube layers capucine orange, orange to argus brown, woody hard, not stratified, or indistinct, up to 4.9 cm thick. Pores surface shining, raw sienna to antique brown when fresh, orange, argus brown to medal bronze when dry, sterile margin absent or narrow. Pores surface of old basidiocarps (4–5 years) orange citrine to medal bronze; sterile margin narrow to 2.8 mm width. Pores circular to angular, 5–6 per mm; dissepiments thin to thick, entire or some lacerate on margin.
Hyphal system dimitic, generative hyphae simple septate, tissue darkening in KOH, unchanged in Melzer’s reagent.
Generative hyphae from tomentum hyaline, thin-walled, frequently branched with simple septate, 1.6–3.8 µm in diam. Skeletal hyphae from tomentum yellow to brown, thick-walled with a wide to narrow lumen, occasionally branched, simple septate often in part of hyphae with wide lumen, 4.6–14.1 µm in diam.
Context generative hyphae hyaline to pale yellow, thin- to slightly thick-walled, frequently branched, simple septate, 2.6–5.2 µm in diam; context skeletal hyphae dominant, orange to brown, occasionally branched, thick-walled with a wide to narrow lumen, simple septate often in part of hyphae with wide lumen, 3.6–10.6 µm in diam.
Tramal generative hyphae hyaline to pale yellow, thin- to slightly thick-walled, simple septate, frequently branched, 2.4–3.7 µm in diam; tramal skeletal hyphae orange to brown, dominant, rare branched, thick-walled with a wide to narrow lumen, simple septate occasionally in part of hyphae with wide lumen, 3.1–8.2 µm in diam.
Hymenial setae lacking; cystidioles present, fusoid, hyaline, thin-walled, 9.9–15.4 × 2.9–3.5 µm; basidia clavate to barrel-shaped, hyaline, with basal simple septum and four sterigmata, 7.9–17.4 × 2.8–6.8 µm; basidioles clavate, barrel to elliptical shape, 8.3–18.8 × 3–6.8 µm.
Basidiospores broadly ellipsoid, brown, slightly thick-walled, smooth, IKI−, CB−, or weak reaction (4.2−)4.3 − 5.3(−5.5) × (3.1−)3.3 − 4.2 µm, L = 4.93 µm, W = 3.73 µm, Q = 1.32 (n = 62).
Habitat: growing on living angiosperm trunks.
Additional specimens examined: China, Guangxi, Chongzuo, Nonggang Nature Reserve, on living angiosperm trunks, June 20 2012, GXU0501, GXU0766, and GXU1102; on dead angiosperm trunks, November 18 2018, GXU2254.
Note: Differs from other species by basidiocarps perennial, sessile, solitary, pileus applanate, the periphery of pilei radially cracked on old fruiting body. Pores circular to angular, 5–6 per mm. Hyphal system dimitic, setae absent, cystidioles fusoid, 9.9–15.4 × 2.9–3.5 µm, basidiospores broadly ellipsoid, 4.93 × 3.73 µm on average.
Fulvifomes tubogeneratus F.C. Huang, H.F. Zheng & Bin Liu, sp. nov. (Figures 5 and 6).
Figure 5.
Basidiocarps of Fulvifomes tubogeneratus. A: basidiocarps imbricate; B: new fruit bodies generated from tube surface. Scales bar: 1 cm.
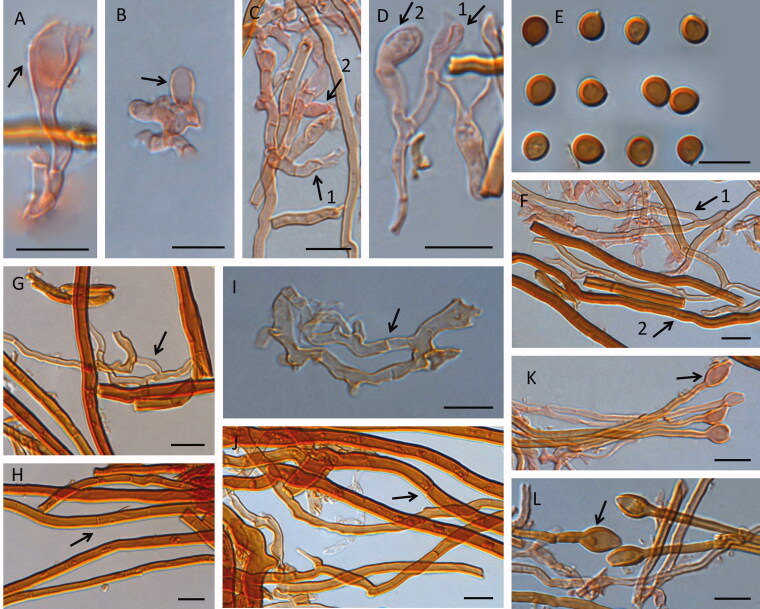
Figure 6.
Microscopic structures of Fulvifomes tubogeneratus. Scales bar: 10 μm. A: basidia; B: basidioles; C, D: 1, basidia, 2, basidioles; E: basidiospores; F: 1, tramal generative hyphae, 2, tramal skeletal hyphae; G: generative hyphae from upper context (arrow pointed); H: skeletal hyphae from upper context (arrow pointed); I: generative hyphae from lower context; J: skeletal hyphae from lower context (arrow pointed); K, L: hyphoid setae swollen at apex from tramal.
MycoBank: MB835791
Etymology: tubogeneratus (Lat.): referring to new fruit body generated from tubes surface.
Type: China, Guangxi Autonomous Region, Chongzuo, Longzhou County, Nonggang Nature Reserve, on dead trunks of angiosperm tree, November 19 2018, GXU2468 (Holotype in GXU).
rDNA sequences ex holotype: MT580805 (ITS), MT580800 (nLSU).
Description: Basidiocarps perennial, sessile, broadly attached on dead drunk, frequently new fruit bodies generated on surface of tubes, occasionally on pileal surface, imbricate. Pileus semicircular to subcircular, applanate, up to 10.2 × 7.4 and 4.1 cm thick, pileus surface distinctly concentrically sulcate, velutinate, brussels brown to medal bronze, margin entire, acute or dull, cadmium yellow to raw sienna, and claret brown when age. Context wood hard, yellow ocher, buckthorn brown to cinnamon brown, duplex, separated by a black line, the upper context up to 0.6 cm, lower context up to 3.1 cm, black lines also frequently distributing in context, tubes, and between context and tube with age. Pore surface shining, rood’s brown to burnt umber, pores not appearing on too young fruit body, but gradually increasing with mature, and some places of surface no pores differentiation and development even fruit body becoming old, and remaining with velutinate, these places capucine yellow, orange to cadmium yellow when young, brussels brown when old, sterile margin with the same color, up to 2.2 cm (no pores differentiation area) even old, pores angular or circular, 7–8 per mm; dissepiments most thin, and parts thick, entire, or some lacerate on margin. Tubes concolorous with pore surface, woody hard, up to 0.8 cm long, not stratified when young, but indistinct stratified with age, rood’s brown to chocolate.
Hyphal system dimitic; generative hyphae simple septate; tissue darkening in KOH, unchanged in Melzer’s reagent.
Upper context generative hyphae frequently branched, simple septate, hyaline to pale yellow, thin to slightly thick-walled with a wide lumen, 1.7–3.3 µm in diam; skeletal hyphae dominant, unbranched, capucine orange to orange, mostly thick-walled with a narrow to wide lumen and septate, and some still with a part of solid, 3–6 µm in diam. Lower context generative hyphae hyaline to pale yellow, frequently branched, thin to slightly thick-walled with a wide lumen, simple septate, 1.9–6.1 µm in diam; skeletal hyphae frequent, occasionally branched, capucine orange to orange, thick-walled with a narrow to wide lumen, a few septate, 3.6–7.2 µm in diam.
Tramal generative hyphae frequent, hyaline to pale yellow, thin to slightly thick-walled, frequently branched, septate, 1.7–3.8 µm in diam; skeletal hyphae frequent, thick-walled with wide to narrow lumen, or solid, occasionally branched, rare septate, pale yellow to orange, 2.5–4.7 µm in diam.
Hymenial setae absent; hyphoid setae pale yellow to orange, frequent swollen at apex, and subglobose to fusiform, thick-walled, 7–19.8 × 4–7.2 µm; basidia clavate to barrel-shaped, flask-shaped, with four sterigmata, and a simple septum at the base, 9.9–31.4 × 3.1–7.6 µm; basidioles clavate to barrel-shaped, subglobose, 7.5–27.5 × 2.8–8 µm; cystidia and cystidioles absent.
Basidiospores ovoid to subglobose, capucine yellow to mars yellow, slightly thick-walled, smooth, IKI−, CB−, (5−)5.2 − 6.2(−6.4) × (4.1−) 4.5 − 5.7(−5.8) µm, L = 5.72 µm, W = 5.00 µm, Q = 1.14 (n = 60).
Habitat: growing on dead trunks of angiosperm trees.
Additional specimens examined: China, Guangxi, Chongzuo, Nonggang Nature Reserve, on dead angiosperm trunks, November 20 2018, GXU2478.
Note: Differs from other species by basidiocarps perennial, sessile, imbricate, new fruit bodies frequently generated from surface of tubes, context duplex, pores angular or circular, 7–8 per mm. Hyphal system dimitic, hyphoid setae present, basidiospores ovoid to subglobose, 5.72 × 5.00 µm on average.
4. Discussion
Morphology and DNA sequence analysis confirmed that the unique of the two new species F. nonggangensis and F. tubogeneratus. Species in the genus Fulvifomes are somewhat heterogeneous in certain characters, such as the presence or absence of setae and spores being cyanophilous (CB+) or acyanophilous (CB–) [2]. However, the basidiospores of two new species in this investigation were both CB–, and hymenial setae absent as well.
Compared with other species, especially querying keys built by Dai [2] and Hattori et al. [11], F. indicus (Massee) L.W. Zhou, F. merrillii (Murrill) Baltazar & Gibertoni, and F. squamosus Salvador-Montoya & Drechsler-Santos are similar to F. nonggangensis and F. tubogeneratus in morphology, all of their basidocarps pileate, applanate, or ungulate to applanate, tube layers not stratified to indistinct, hymenial setae absent. Nevertheless, each of them has some specific characteristics different from two new species.
F. indicus is distinguished from two new species by basidiocarps annual [10], sessile or substipitate with a contracted base, hyphal system monomitic, pores the largest (4–5 per mm). Furthermore, basidiospores is larger (5.4–6.5 × 4.7–5.5 µm) than that of F. nonggangensis.
According to the data of Dai [2], F. merrillii differs from F. nonggangensis by pilei subungulate to applanate, pores smaller (7 − 8 per mm), tube layers indistinct, cystidioles larger (18–22 × 4.5–6 µm), basidiospores subglobose; and differs from F. tubogeneratus by basidiocarps subungulate to applanate, solitary, cystidioles present, and hyphoid setae absent and basidiospores smaller (4.4–5.4 × 3.7–4.7 µm).
F. squamosus [13] is distinguishable from two new species by having squamose pilear surface with long scales, hyphal system monomitic in the context, basidiospores with the ventral side flattened, and without cystidioles and hyphoid setae.
Similar to previous phylogenetic studies on Fulvifomes [12,14,15], the core of Fulvifomes in phylogenetic trees was mainly divided into two clades, one including F. fastuosus, F. imbricatus, F. siamensis et al., another including F. elaeodendri, F. krugiodendri, F. squamosus, F. thailandicus, and F. yoroui et al.
F. nonggangensis is closely related to F. XHJ-2018b, F. rhytiphloeus (Mont.) Camp.-Sant. & Robledo, and I. rigidus B.K. Cui & Y.C. Dai, meanwhile, F. tubogeneratus closely related to F. XHJ-2018h and F. siamensis T. Hatt., Sakay. & E.B.G. Jones (Figures 1 and 2), isolates of F. XHJ-2018b and F. XHJ-2018h both came from China but were not formally described yet.
F. rhytiphloeus was proposed by Campos-Santana et al. [26] as a new complex species, and it resembles F. nonggangensis by basidiocarps pileate, applanate, solitary, with a distinct black line below pileus surface, setae absent. But its tubes were mostly distinctly stratified, pores smaller, 7–9 per mm, context fibrous and easily fragmented, cystidioles absent, and spores subglobose.
I. rigidus [27] differed from F. nonggangensis in its basidiocarps annual, resupinate, pores smaller, 8–9 per mm, hyphal system monomitic, cystidioles absent, and slightly smaller basidiospores (3.9–4.5 × 2.9–3.7 µm). But it also fitted in Fulvifomes with hymenial setae absent, ellipsoid, yellowish brown and thick-walled basidiospores, whether it is necessary to transferred I. rigidus to Fulvifomes which still needed more evidences.
F. siamensis was proposed by Hattori et al. [11], and resembled F. tubogeneratus by basidiocarps perennial, context woody hard, pores 7–8 per mm, setae absent. But it differed from the latter by without a distinct black line, and hyphal system monomitic in context, hyphoid setae absent. Moreover, basidiocarps of F. tubogeneratus imbricate, and new fruit bodies often generated from tube surface.
Key to species of Fulvifomes from China [2,10,28]
1.Hymenial setae absent2
1.Hymenial setae present11
2.Basidiocarps resupinate3
2.Basidiocarps pileate, effused-reflexed4
3.Basidiospores larger, 4.3–5.1 × 3.4–4.2 µm, cystidioles presentF. inermis
3.Basidiospores smaller, 3.1-4.2 × 2.6-3.1 µm, cystidioles absentF. membranaceus
4.Basidiocarps effused-reflexed5
4.Basidiocarps pileate6
5.Tube layer distinct, basidiospores ellipsoid, 4.7–5.8 × 3.7–4.6 µmF. macgregorii
5.Tube layer not stratified, basidiospores oblong-ellipsoid, 4.2–5.1 × 3–3.5 µmF. collinus 6. Basidiocarps annual, hyphal system monomiticF. indicus
6.Basidiocarps perennial, hyphal system dimitic7
7.Tube layer distinct8
7.Tube layer not stratified to indistinct9
8.Pores 3–4 per mm, spores ellipsoid, chlamydospores absentF. hainanensis
8.Pores 6–7 per mm, spores subglobose, chlamydospores presentF. durissimus
9.Hyphoid setae present, cystidioles absent F. tubogeneratus
9.Hyphoid setae absent, cystidioles present10
10.Pores 5–6 per mm, spores broadly ellipsoid, 4.3 − 5.3 × 3.3 − 4.2 µmF. nonggangensis
10.Pores 7–8 per mm, spores subglobose, 4.4–5.4 × 3.7–4.7 µmF. merrillii
11.Basidiocarps effused-reflexed to pileate, pores 6–7 per mmF. johnsonianus
11.Basidiocarps resupinate, pores 8–11 per mm12
12.Basidiospores 2–2.5 µm long, tube layers distinctF. minisporus
12.Basidiospores 2.3–4.1 µm long, tube layers not stratified to indistinct13
13.Basidiocarps annual, pore surface cracked when dry, spores CB(−)F. glaucescens
13.Basidiocarps perennial, pore surface not cracked when dry, spores CB(+)F. cesatii
Funding Statement
This study was supported by National Natural Science Foundation of China [No. 3196006], the Key Research and Development Plan of Guangxi, China [No. AB18221047], and Project of Guangxi Innovation Team of National Modern Agricultural Industry System [nycytxgxcxtd-07-01].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Ediriweera SS, Wijesundera RLC, Nanayakkara CM, et al. . A new record of Fulvifomes fastuosus recorded from Sri Lanka. J Natn Sci Foundation Sri Lanka. 2014;42(4):369–377. [Google Scholar]
- 2.Dai YC. Hymenochaetaceae (Basidiomycota) in China. Fungal Divers. 2010;45(1):131–343. [Google Scholar]
- 3.Wu F, Zhou LW, Yang ZL, et al. . Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species. Fungal Divers. 2019;98(1):1–76. [Google Scholar]
- 4.Murrill WA. Northern polypores. New York (NY): Privately Printed; 1914. [Google Scholar]
- 5.Ryvarden L, Johansen I.. A preliminary polypore flora of East Africa. Oslo, Norway: Fungiflora; 1980. [Google Scholar]
- 6.Gilbertson RL, Ryvarden L.. North American polypores. Oslo, Norway: Fungifora; 1987. [Google Scholar]
- 7.Ryvarden L, Gilbertson RL. European polypores. Vol. 2. Synopsis fungorum. Oslo (Norway): Fungiflora press; 1993. [Google Scholar]
- 8.Nunez M, Ryvarden L. East Asian polypores. Vol. 1. Ganodermataceae and Hymenochaetaceae. Synopsis Fungorum. Oslo (Norway): Fungiflora press; 2000. [Google Scholar]
- 9.Wagner T, Fischer M.. Proceedings towards a natural classification of the worldwide taxa Phellinus s.l. and Inonotus s.l., and phylogenetic relationships of allied genera. Mycologia. 2002;94:998–1016. [PubMed] [Google Scholar]
- 10.Zhou LW. Fulvifomes hainanensis sp. nov. and F. indicus comb. nov. (Hymenochaetales, Basidiomycota) evidenced by a combination of morphology and phylogeny. Mycoscience. 2014;55(1):70–77. [Google Scholar]
- 11.Hattori T, Sakayaroj J, Jones EBG, et al. . Three species of Fulvifomes (Basidiomycota, Hymenochaetales) associated with rots on mangrove tree Xylocarpus granatum in Thailand. Mycoscience. 2014;55(5):344–354. [Google Scholar]
- 12.Ji XH, Dai YC, Vlasák J.. Two new species of Fulvifomes (Hymenochaetales, Basidiomycota) from America. MycoKeys. 2017;22:1–13. [Google Scholar]
- 13.Salvador-Montoya CA, Popoff OF, Reck M, et al. . Taxonomic delimitation of Fulvifomes robiniae (Hymenochaetales, Basidiomycota) and related species in America: F. squamosus sp. nov. Plant Syst Evol. 2018;304(3):445–459. [Google Scholar]
- 14.Olou BA, Ordynets A, Langer E.. First new species of Fulvifomes (Hymenochaetales, Basidiomycota) from tropical Africa. Mycol Prog. 2019;18(12):1383–1393. [Google Scholar]
- 15.Tchoumi JMT, Coetzee MPA, Rajchenberg M, et al. . Poroid Hymenochaetaceae associated with trees showing wood-rot symptoms in the Garden Route National Park of South Africa. Mycologia. 2020;112:722–741. [DOI] [PubMed] [Google Scholar]
- 16.Ridgway R. Color standards and color nomenclature. Washington (DC): United States National Museum; 1912. [Google Scholar]
- 17.White TJ, Bruns T, Lee S, et al. . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols, a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322. [Google Scholar]
- 18.Vilgalys R, Hester M.. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, et al. . MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewnlak F, et al. . The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford DL. PAUP*, phylogenetic analysis using parsimony, ver. 4.0b10. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- 22.Nylander JAA. MrModeltest v2. Evolutionary biology center. Uppsala, Sweden: Uppsala University; 2004. [Google Scholar]
- 23.Posada D, Crandall KA.. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F, Teslenko M, P van der M, et al. . MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stöver BC, Müller KF.. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 2010;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campos-Santana M, Robledo G, Decock C, et al. . Diversity of the poroid Hymenochaetaceae (Basidiomycota) from the Atlantic Forest and Pampa in Southern Brazil. Cryptogam Mycol. 2015;36(1):43–78. [Google Scholar]
- 27.Cui BK, Du P, Dai YC.. Three new species of Inonotus (Basidiomycota, Hymenochaetaceae) from China. Mycol Prog. 2011;10(1):107–114. [Google Scholar]
- 28.Wang K, Zhao MJ, Su JH, et al. . The use of Checklist of Fungi in China database in the red list assessment of macrofungi in China. Biodivers Sci. 2020;28(1):74–98. [Google Scholar]