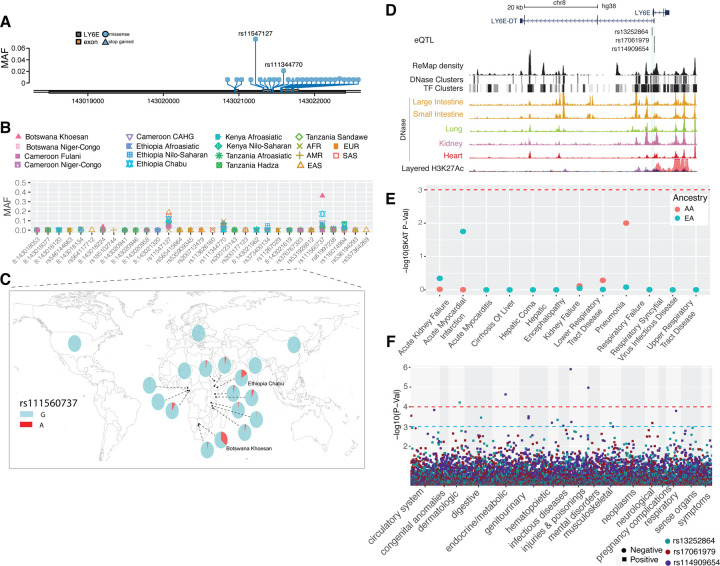
Figure 7. Genetic variation at LY6E and its disease association.
(A) Location of coding variants and their minor allele frequency (MAF) at LY6E identified from the pooled dataset. (B) MAF of coding variants in diverse global ethnic groups. (C) The MAF of variant rs111560737 at LY6E in diverse global ethnic groups. Each pie denotes frequencies of alleles in the corresponding population. (D) Three regulatory eQTLs identified at LY6E. RNA Pol2 ChIA-PET data and DNase-seq data of large intestine, small intestine, lung, kidney and heart are from ENCODE81. (E) Gene-based association result between coding variants at LY6E and 12 disease classes. The disease classes are shown on the x-axis and the y-axis represents the p-values. EA, European Ancestry; AA, African American ancestry. (F) PheWAS plot of the three eQTL associated with LY6E and ~1800 disease codes across 17 disease categories. The disease categories are shown on the x-axis and the y-axis represents the −log10 of the p-values. The colored dot represents an eQTL and the direction of effect of the association. The red dashed line denotes the 0.0001 cutoff, and the blue dashed line represents the 0.001 cutoff.