Abstract
Legume and rhizobium species can establish a nitrogen-fixing nodule symbiosis. Previous studies have shown that several transcription factors that play a role in (lateral) root development are also involved in nodule development. Chromatin remodeling factors, like transcription factors, are key players in regulating gene expression. However, studies have not investigated whether chromatin remodeling genes that are essential for root development are also involved in nodule development. Here, we studied the role of Medicago (Medicago truncatula) histone deacetylases (MtHDTs) in nodule development. Arabidopsis (Arabidopsis thaliana) orthologs of HDTs have been shown to play a role in root development. MtHDT expression is induced in nodule primordia and is maintained in the nodule meristem and infection zone. Conditional, nodule-specific knockdown of MtHDT expression by RNAi blocks nodule primordium development. A few nodules may still form, but their nodule meristems are smaller, and rhizobial colonization of the cells derived from the meristem is markedly reduced. Although the HDTs are expressed during nodule and root development, transcriptome analyses indicate that HDTs control the development of each organ in a different manner. During nodule development, the MtHDTs positively regulate 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 (MtHMGR1). Decreased expression of MtHMGR1 is sufficient to explain the inhibition of primordium formation.
Plant-specific histone deacetylases regulate the expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductases to control root nodule development.
Introduction
Plants are able to develop lateral organs post-embryonically. An example is the formation of lateral roots (Malamy and Benfey, 1997). Roots of legume plants have the ability to form a second lateral organ, root nodules. The latter are symbiotic organs, which are used to host rhizobium bacteria. These become able to reduce atmospheric nitrogen into ammonia, which can be used by the host (Udvardi and Poole, 2013).The model legume Medicago (Medicago truncatula) forms indeterminate nodules. Their histology and ontology bear resemblance to that of (lateral) roots. In both organs, a meristem is present at their apex (Franssen et al., 1992; van den Berg et al., 1995), which is followed by a zone containing differentiating cells. This is the elongation zone in roots and the infection zone in nodules (Vinardell et al., 2003; Vanstraelen et al., 2009). In the latter, intracellular infection by rhizobia takes place. The fully differentiated cells form the differentiated zone in roots and the fixation zone in nodules. The switch from infection to fixation zone is characterized by the sudden accumulation of starch in the infected cells (Gavrin et al., 2014). In Medicago, both nodules and lateral roots are developed from primordia, whose formation is initiated at the protoxylem pole and starts with cell division in pericycle, and subsequently, divisions are induced in endodermis and cortex (Dubrovsky et al., 2001; Xiao et al., 2014, 2019). Therefore, nodules and lateral roots exhibit similarities in organogenesis.
Recent studies showed that some transcription factors involved in (lateral) root development have been recruited for nodule development. In Medicago, knockdown of PLETHORA genes known to be key regulators in root development, blocks nodule meristem activity (Aida et al., 2004; Franssen et al., 2015), and knockout of LOB-DOMAIN PROTEIN 16 (LBD16) reduces both nodule and lateral root initiation (Goh et al., 2012; Schiessl et al., 2019). It is known that chromatin remodeling factors contribute to transcriptional reprogramming and also play a central role in plant organ development (Jarillo et al., 2009). However, whether chromatin remodeling factors, which are involved in root development, also have a role in nodule development has never been studied.
Some of the best studied chromatin remodeling factors are histone deacetylases (HDACs), which are responsible for inactivation of genes. They can be grouped into three different classes, reduced potassium dependency 3 (RPD3), silent information regulator (SIR2), and HDT (Pandey et al., 2002). The last group, HDT, is plant-specific and has no homology to HDACs in animals and fungi (Lusser et al., 1997). Previously, we have shown that in Arabidopsis, two HDTs (AtHDT1/2) are expressed in the root meristem, and control its size by repressing C19-GIBBERELLIN 2-OXIDASE 2 (AtGA2ox2; Li et al., 2017). Further, AtHDTs are markedly upregulated in dedifferentiating pericycle cells during the initiation of lateral root primordia (De Smet et al., 2008). Medicago contains three HDT members; Medtr4g055440, Medtr2g084815, and Medtr8g069135, which were designated as MtHDT1, MtHDT2, and MtHDT3, respectively (Grandperret et al., 2014). Laser capture microdissection RNA sequencing analyses indicated that they all are expressed in nodule meristem and infection zone (Roux et al., 2014). Here, we studied whether Medicago HDTs play a role in nodule development, and if so, whether they have a similar function as in the root development.
We showed that the three MtHDTs are expressed in young nodule primordia. In mature nodules they are expressed in the meristem and the infection zone. Knockdown of MtHDTs in a nodule specific way (ENOD12::MtHDTs RNAi) blocks cell division in most of the nodule primordia. In the few nodules formed on RNAi transgenic roots, meristem size and activity, as well as rhizobial colonization, are reduced. Transcriptome analysis of RNAi nodules showed that HDTs regulate nodule and root development in a different manner. The differentially expressed genes (DEGs) in RNAi nodule primordia and in mature nodules in part overlap, and in both cases, expression of the MtHMGR1 is reduced.
Results
Medicago HDT2 has a similar function as Arabidopsis HDT1/2 in controlling root development
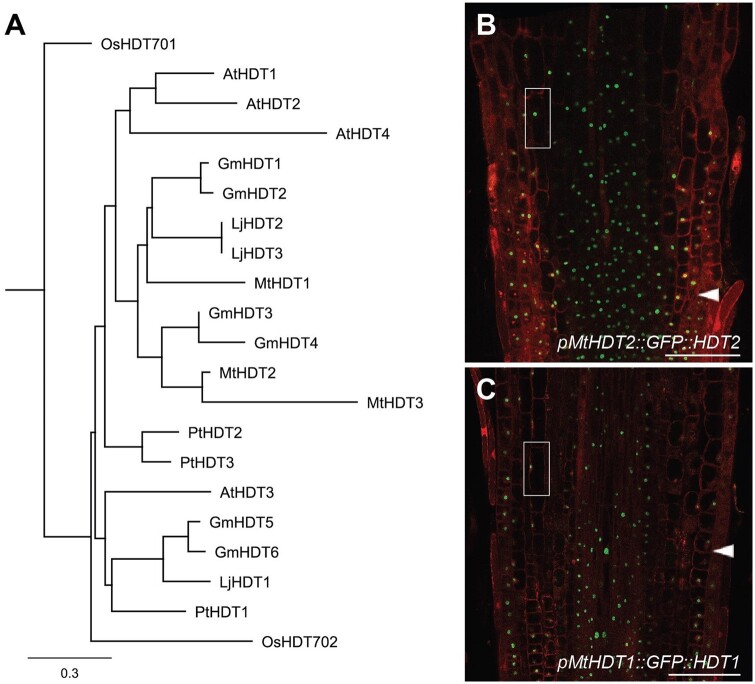
To compare the functions of the Medicago HDTs with the previously characterized Arabidopsis HDTs, we first analyzed the phylogenetic relationship of HDTs by using protein sequences from several dicots and the monocot rice. This showed that HDTs in rice were divergent from those in dicots. Within dicots, HDTs have evolved into two clades (Figure 1, A andSupplemental Table S1). The first clade contained the Arabidopsis AtHDT3 and none of the Medicago MtHDTs. The second clade contained AtHDT1, 2, 4, and all three MtHDTs. Further, independent duplications have occurred in the three legume species, Medicago, Lotus, and Soybean, and this has resulted in high sequence similarity of HDT pairs. In Medicago, such a pair is formed by MtHDT2 and 3. In Arabidopsis, a similar independent duplication resulted in AtHDT1 and 2. Previously, we showed that AtHDT1 and 2 are functionally redundant and are essential for root growth. AtHDT4 regulates root growth as well (Han et al., 2016). Therefore, it is very likely that some of the MtHDTs are involved in root development.
Figure 1.
MtHDTs are homologous to AtHDT1, 2. A, Phylogenetic tree of HDT proteins. The protein sequences are obtained from Medicago truncatula (Mt), Lotus japonicus (Lj), Glycine max (Gm), Arabidopsis thaliana (At), Populus trichocarpa (Pt), and Oryza sativa (Os). Scale bar represents substitutions per site. B and C, Localization of pMtHDT2::GFP::HDT2 (B) and pMtHDT1::GFP::HDT1 (C) in longitudinal sections of Medicago root tips. Magnifications of boxed areas are shown in Supplemental Figure S1. Arrowheads indicate the boundary between root meristem and elongation zone. Scale bars = 100 µm.
To study whether MtHDTs have a similar expression pattern as AtHDTs in roots, we generated GFP-MtHDT constructs including a ∼2-kb DNA region upstream of the start codon (putative promoter), GFP, and the corresponding MtHDT coding sequence (pMtHDT1::GFP::HDT1, pMtHDT2::GFP::HDT2, and pMtHDT3::GFP::HDT3). These constructs were introduced into Medicago by Agrobacterium rhizogenes mediated hairy root transformation (Limpens et al., 2004). In Medicago roots, pMtHDT1::GFP::HDT1 and pMtHDT2::GFP::HDT2 were expressed in the meristem and elongation zone and localized to the nucleus. Propidium iodide stained the whole nucleus in fixed material, while GFP-MtHDTs were mainly restricted within a round compartment in the nucleus, indicating MtHDT proteins are mainly localized in the nucleoli (Figure 1, B and C and Supplemental Figure S1, A). In the differentiated zone, these fusion proteins were hardly detected. This is similar to the expression pattern and the subcellular localization of AtHDT1 and AtHDT2 in Arabidopsis root tips (Li et al., 2017). Expression level of MtHDT2 in root tips was higher than that of MtHDT1. Expression of pMtHDT3::GFP::HDT3 was below detection level, and therefore we studied the MtHDT3 expression pattern using a pMtHDT3::GUS construct that included the putative MtHDT3 promoter and β-glucuronidase (GUS) coding sequence. The construct was introduced into Medicago by hairy root transformation and it demonstrated that pMtHDT3::GUS was weakly expressed in the root meristem (Supplemental Figure S2).
The high sequence similarity and the similar expression pattern of HDTs in Arabidopsis and Medicago roots suggest that they may control root growth in the same way. Mthdt2 mutants containing Tnt1 insertions either in the third exon or in the eighth intron have recently become available, but they have a wild-type-like root phenotype (Supplemental Figure S3), and for the other HDT genes, mutants are not available. To test whether MtHDTs play a role in Medicago root growth, we created a RNA interference construct driven by the CaMV 35S promoter to target all three MtHDTs transcripts (35S::MtHDTs RNAi). The 35S promoter is known to be active in Medicago root tips (Samac et al., 2004), and this matches MtHDTs expression pattern. In 35S::MtHDTs RNAi root tips, expression of MtHDT1, 2, and 3 was knockeddown to 17%, 16%, and 33% of the control level, respectively. Their root meristem size was ∼14% significantly reduced (Supplemental Figure S4), similar to that in Arabidopsis AtHDT1, 2 knockdown roots. To determine which MtHDT gene is sufficient to support root growth in Arabidopsis, we introduced each pMtHDT::GFP::HDT construct into a double heterozygous HDT1hdt1 HDT2hdt2 Arabidopsis mutant. Loss of function of both AtHDT1 and AtHDT2 is lethal (Li et al., 2017), and therefore we tested in the progeny of the transformed HDT1hdt1HDT2hdt2 plants which MtHDT gene was able to rescue the lethal phenotype. More than 200 transformed plantlets of each progeny were genotyped, which showed that pMtHDT2::GFP::HDT2 complemented Arabidopsis hdt1hdt1hdt2hdt2, whereas pMtHDT1::GFP::HDT1 and pMtHDT3::GFP::HDT3 did not. Further, in Arabidopsis roots, pMtHDT2::GFP::HDT2 was expressed in the meristem and elongation zone and mainly localized in nucleoli (Supplemental Figure S5), similar to AtHDT1/2 (Li et al., 2017). Together, the expression pattern studies and complementation test suggest that MtHDT2 has a similar role as AtHDT1, 2 in root development. It does not exclude that MtHDT1 and 3 are also involved in root development, as they are expressed in Medicago root tips.
MtHDTs are expressed in the nodule meristem and infection zone
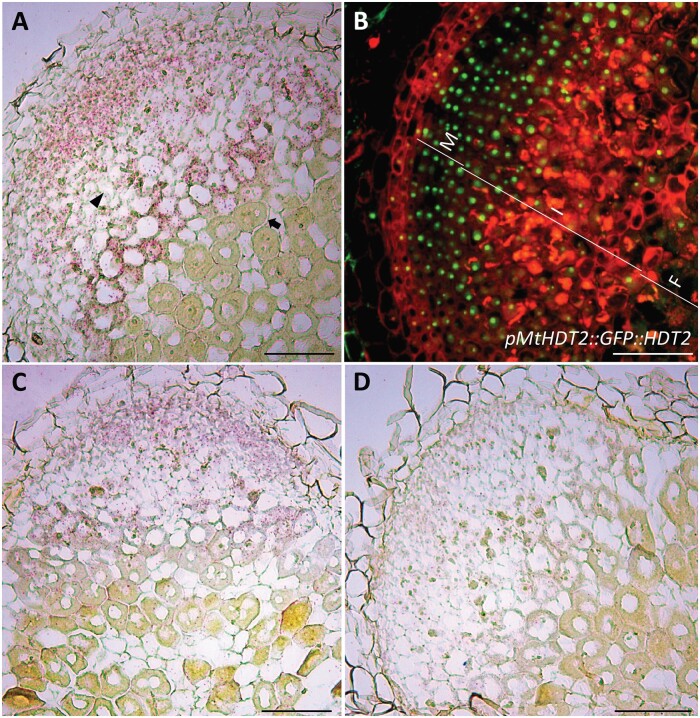
In this study, we especially focused on the role of MtHDTs in nodule development. As all three Medicago HDTs are expressed in roots, and nodule and root development are related, we first studied all three Medicago genes. To determine where MtHDTs are expressed in nodules, we performed RNA in situ hybridization on longitudinal sections of nodules, using probe sets specific for each MtHDT. We used in situ hybridization as this gives the most accurate expression pattern, especially since we could not test in Medicago whether the selected MtHDT promoter regions are biologically functional. The in situ hybridization experiment showed that MtHDT2 transcripts were present at a similar level in both the nodule meristem and infection zone (Figure 2, A). In the latter, MtHDT2 was mainly expressed in infected cells and hardly detectable in uninfected cells. This is different from roots, in which HDT genes are only expressed in the meristem (Li et al., 2017). At the transition from infection to fixation zone, the expression of MtHDT2 dropped dramatically. The spatial distributions of MtHDT1 and MtHDT3 transcripts were similar to that of MtHDT2, but the hybridization signals were markedly lower (Figure 2, C and D). So like in roots, MtHDT2 is more highly expressed in nodules than the other MtHDTs. In addition, MtHDT2 is certainly involved in root development. Therefore, in further experiments we focused on this gene.
Figure 2.
MtHDTs are expressed in the nodule meristem and infection zone. A, Expression of MtHDT2 mRNA visualized by in situ hybridization in wild-type Medicago nodules. The arrowhead indicates a noninfected cell in the infection zone and the arrow indicates a cell of the first cell layer of the fixation zone where amyloplasts are detectable at the periphery. B, Localization pattern of pMtHDT2::GFP::HDT2 in nodules. The nodule meristem zone (M), infection zone (I), and fixation zone (F) are marked. C and D, Expression of MtHDT1 (C) and MtHDT3 (D) mRNA visualized by in situ hybridization in wild-type Medicago nodules. Images are longitudinal sections of nodules harvested at 21 dpi. Representative images are shown. In A, C, and D, red dots are hybridization signals. A–D, Scale bars = 100 µm.
To determine the subcellular localization of MtHDT2 protein in nodules, we used the pMtHDT2::GFP::HDT2 construct. This showed that MtHDT2 protein accumulated in cells of nodule meristem and infection zone, and like in roots, mainly in nucleoli (Figure 2, B andSupplemental Figure S1, B). Further, at the switch from infection to fixation zone, MtHDT2 protein level suddenly dropped to below detection level. So, the distribution of the protein is similar to that of the transcript. Further, the expression of MtHDT2 in nodule meristem and infected cells of the infection zone indicated that this gene might control meristem activity, rhizobial release, and/or intracellular accommodation of rhizobia.
Meristem activity and rhizobial colonization require MtHDTs
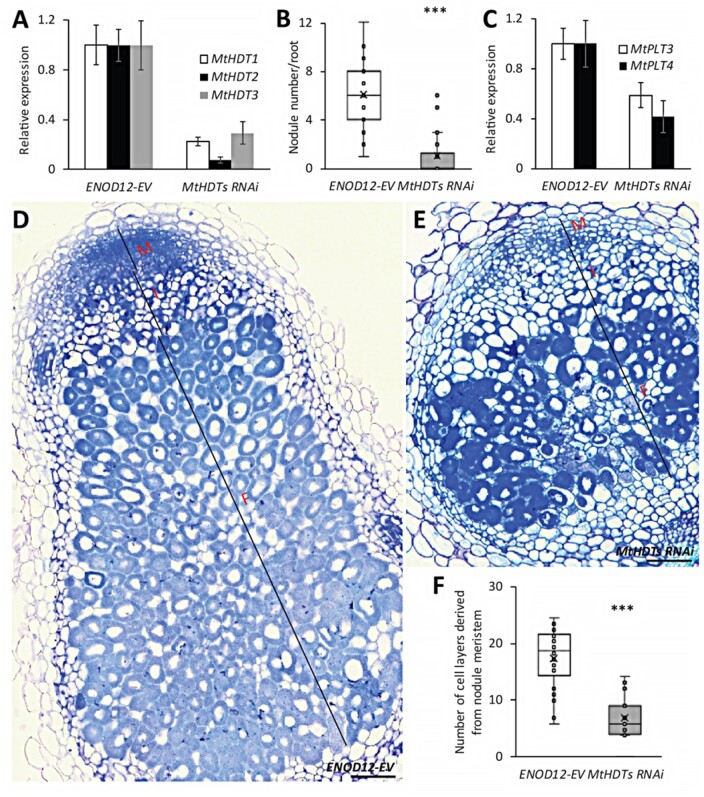
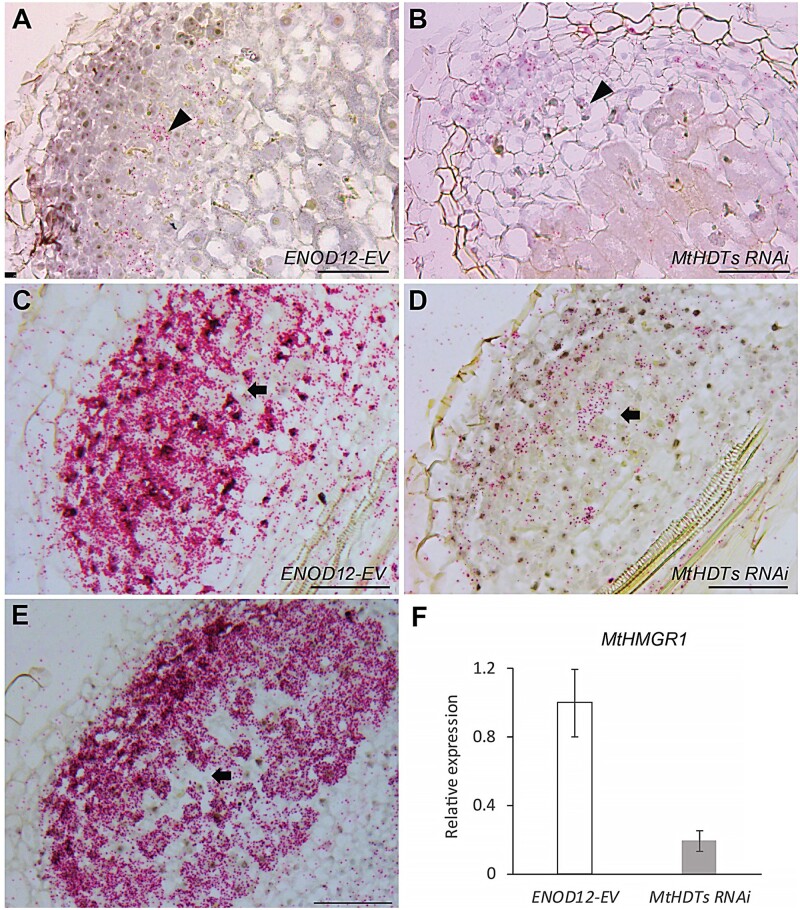
To determine the role of MtHDTs in nodules, we made a nodule-specific RNA interference construct to target all three MtHDT transcripts (ENOD12::MtHDTs RNAi). Although MtHDT2 has the highest expression level in nodules, we decided to also knockdown the other two MtHDTs, as a MtHDT2 Tnt1 mutant has no nodule phenotype (Supplemental Figure S3). We used the ENOD12 promoter to drive the RNAi construct as it is active in the nodule meristem and infection zone, and it therefore covers the expression domains of the three MtHDTs (Limpens et al., 2005; Franssen et al., 2015). In the RNAi transgenic nodules, MtHDT1, 2, and 3 were knockeddown to 22%, 7%, and 29% of the levels in ENOD12-EV (Empty Vector) control nodules, respectively (Figure 3, A). At 21 d post inoculation (dpi), control roots formed on average 6.0 nodules/root, whereas MtHDTs RNAi roots exhibited only 1.1 nodules/root (Figure 3, B). Although the RNAi nodule number was low, it still allowed for histological characterization.
Figure 3.
Knockdown of MtHDTs affects nodule meristem functioning and rhizobial colonization. A, Reverse transcription quantitative PCR (RT-qPCR) analysis of MtHDTs expression in ENOD12-EV control and MtHDTs RNAi nodules. B, Nodule number formed per ENOD12-EV and MtHDTs RNAi transgenic root (n > 20 roots). C, RT-qPCR analysis of MtPLT3, 4 expression in ENOD12-EV control, and MtHDTs RNAi nodules. D and E, Morphology of ENOD12-EV (D) and MtHDTs RNAi (E) nodules studied by light microscopy. Representative longitudinal sections are shown. The nodule meristem (M), infection zone (I), and fixation zone (F) are marked. Scale bars = 100 µm. Magnifications of nodule infection zone are shown in Supplemental Figure S6. F, Number of cell layers derived from nodule meristem in ENOD12-EV and MtHDTs RNAi transgenic nodules (n > 15). Nodules were harvested at 21 dpi. Panels in A and C show mean ± SEM determined from three independent experiments. Asterisks in B and F indicate significant differences (***P < 0.001; Student’s t test).
The control nodules were elongated, whereas MtHDTs RNAi nodules were spherical and markedly smaller (Figure 3, D and E). Longitudinal sections of control nodules (n = 22) showed that meristems were present at the apex of all nodules and contained approximately eight cell layers (Figure 3, D). Meristems were also present in MtHDTs RNAi nodules (n = 20), but these nodules had only approximately four cell layers (Figure 3, E). In agreement with this reduced number of layers, expressions of MtPLT3 and MtPLT4, two genes that are expressed throughout the nodule meristem (Franssen et al., 2015), were reduced to 59% and 42% of the control level in MtHDTs RNAi nodules (Figure 3, C).
Approximately eight cell layers of the proximal part of the central tissue of a mature nodule are formed and infected at the primordium stage, and these are not derived from the nodule meristem (Xiao et al., 2014). MtHDTs RNAi nodules had about eight cell layers at the proximal part with fully infected cells. These were completely packed with elongated symbiosomes (Figure 3, D and E). This is similar to control nodules. However, the number of cell layers derived from the nodule meristem was markedly reduced (Figure 3, F). Further, in the infected cells in these layers, the colonization level was rather low, resulting in cells with large vacuoles and few bacteria (Supplemental Figure S6). Collectively, these data showed that in the MtHDTs RNAi nodules, knockdown of MtHDTs reduced nodule meristem size, and affected the rhizobial colonization process in cells derived from the meristem, but not from primordium cells.
Knockdown of MtHDTs affects nodule primordium development
As nodule number was markedly reduced on the RNAi roots, we assumed that nodule primordium formation was affected. To test this, we transformed Medicago ENOD11::GUS plants (Boisson-Dernier et al., 2005) with either the MtHDTs RNAi or ENOD12-EV construct, by hairy root transformation. The ENOD11 promoter is active in the whole young nodule primordia, and it is only expressed in one or two cell layers adjacent to root vasculature in lateral root primordia (Supplemental Figure S7). So, it facilitates distinguishing between nodule and lateral root primordia and accurately counting nodule primordium number.
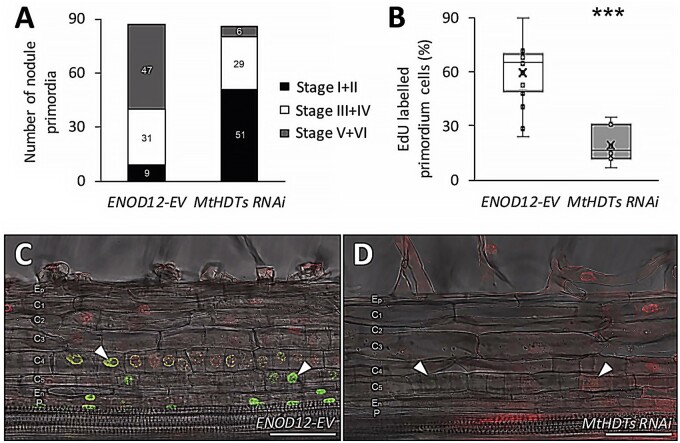
Rhizobia were spot inoculated at the susceptible zones of 110 transgenic ENOD12-EV and 110 MtHDTs RNAi roots with a similar length. After 5 d, 99 control- and 102 MtHDTs RNAi-inoculated roots formed nodule primordia expressing ENOD11. The inoculated root segments with nodule primordia (∼0.3 cm) were embedded in plastic and sectioned to study in which stage nodule primordia had developed. In cases of root segments containing more than one primordium, only the largest nodule primordium was counted. We successfully characterized 87 and 86 control and MtHDTs RNAi segments, respectively. This analysis showed that in control roots, 90% (78 out of 87) of nodule primordia passed stage II, and a relatively high number of them (54%, 47 out of 87) developed into or passed stage V (Figure 4, A). In contrast, on MtHDTs RNAi transgenic roots, the majority of nodule primordia (59%, 51 out of 86) were in stage I or stage II, and only few MtHDTs RNAi nodule primordia (7%, 6 out of 86) had developed into or passed stage V (Figure 4, A). This suggested that the development of the majority of MtHDTs RNAi nodule primordia was blocked at an early stage, which is consistent with reduced nodule number at 21 dpi (Figure 3, B).
Figure 4.
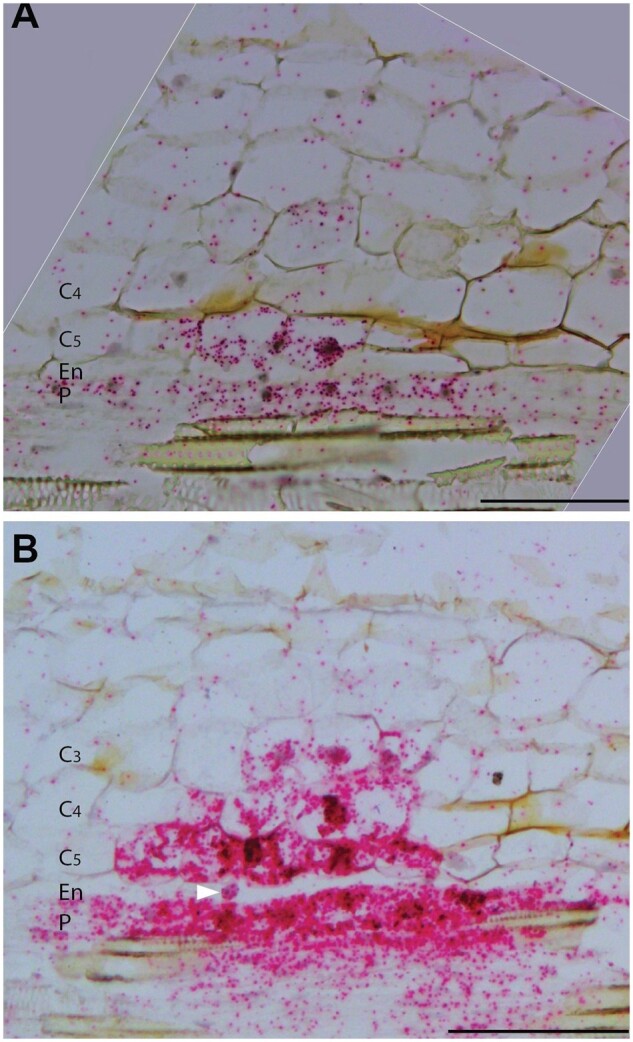
Knockdown of MtHDTs blocks nodule primordium development. A, Analysis of developmental stages of 5 dpi ENOD12-EV (n = 87) and MtHDTs RNAi (n = 86) nodule primordia. Number of primordia at corresponding stages was indicated. B, Percentage of EdU-labeled nodule primordium cells in 2 dpi ENOD12-EV (n = 15) and MtHDTs RNAi (n = 7) nodule primordia. Nodule primordium cells were defined as divided or dividing cells that have a smaller size. Eight MtHDTs RNAi nodule primordia had no EdU labeling and were not used for statistics. Asterisks indicate significant differences (***P < 0.001; Student’s t test). C and D, EdU signals in 2 dpi ENOD12-EV (C) and MtHDTs RNAi (D) nodule primordia. Arrowheads indicate strong (C) or weak (D) green fluorescent signals in nuclei. Red signals are pripordium iodide. Identical confocal microscope settings were used in C and D. P, Pericycle; En, Endodermis; C5/4/3/2/1, the fifth/fourth/third/second/first cortical cell layer; Ep, Epidermis. Scale bars = 100 µm.
To further support that MtHDTs RNAi nodule primordia were blocked in development, root segments containing nodule primordia were collected at 2 d after spot inoculation; they were then incubated for 2 h with EdU, which is incorporated into replicating DNA during mitosis (Kotogany et al., 2010). By quantifying the percentage of EdU-labeled nodule primordium cells, we could determine whether knockdown of MtHDTs reduced mitotic activity in young primordia. Fifteen control and 15 MtHDTs RNAi nodule primordia were analyzed. All control nodule primordia had EdU-labeled cells, and on average, 62% of the primordium cells were labeled (Figure 4, B and C). In contrast, only 47% (7 out of 15) of MtHDTs RNAi nodule primordia had EdU-labeled cells, and in these primordia, the percentage of labeled cells had markedly dropped to 20% (Figure 4, B and D). Further, the intensity of fluorescence in EdU-labeled cells was reduced in comparison with that in control primordia (Figure 4, C and D). Therefore, we concluded that the development of the majority of MtHDTs RNAi nodule primordia had been blocked at the early stages.
MtHDTs are expressed in young nodule primordia
The inhibition of nodule primordium development in MtHDTs RNAi plants prompted us to study whether MtHDTs were expressed in nodule primordia. We first performed RNA in situ hybridization for MtHDT2, as it has the highest expression level, on longitudinal sections of nodule primordia. Cell divisions in Medicago nodule primordia occur first in the pericycle and subsequently in the fifth cortical layer (C5; Xiao et al., 2014). One early stage nodule primordium (stage I) is shown in Figure 5, A, in which MtHDT2 transcripts were present in dividing pericycle and C5 cells. Cell divisions in endodermis are initiated shortly after that, in C3, during nodule primordium development (Xiao et al., 2014). Figure 5, B shows a primordium at stage III, in which cell divisions have occurred in C3, but not yet in the endodermis. However, MtHDT2 transcripts were detected in nuclei of endodermal cells, indicating that MtHDT2 starts to express in cells prior to division.
Figure 5.
MtHDT2 is expressed in nodule primordia. In situ hybridization pattern of MtHDT2 mRNA in nodule primordia at stage I (A) and stage III (B). Longitudinal sections of wild-type nodule primordia are shown. Red dots are hybridization signals. Divided and dividing primordium cells are distinguished by their smaller size (10–60 µm) than nonprimordium cells (>100 µm). Arrowhead in (B) indicates a nucleus from an endodermal cell that has not divided. P, Pericycle; En, Endodermis; C5/4/3, the fifth/fourth/third cortical cell layer. Scale bars = 100µm.
As the expression levels of MtHDT1, 3 are rather low, we were not able to study their expression in primordia with in situ hybridization. Therefore, the expression patterns of MtHDT1 and MtHDT3 in nodule primordia were studied by using promoter-GUS constructs. The pMtHDT1::GUS construct was generated by fusing the MtHDT1 putative promoter with GUS coding region and the pMtHDT3::GUS construct was generated as mentioned previously. These two constructs were introduced into Medicago by hairy root transformation, and transgenic roots were inoculated with rhizobia. We first analyzed their expression patterns in nodules. The GUS expression patterns were consistent with RNA in situ hybridization (Figure 2, C and D and Supplemental Figure S8, A and B), indicating that the putative promoters are sufficient to create the correct gene expression pattern. In nodule primordia, both MtHDT1 and MtHDT3 promoters showed a similar expression pattern as MtHDT2 (Supplemental Figure S8, C and D). The expression of MtHDTs in young primordia indicates that they have a role in nodule primordium initiation and development.
Knockdown of MtHDTs alters gene expression in nodules
HDT proteins are known to regulate chromatin status, thereby contributing to the regulation of gene transcription (Kouzarides, 2007). To investigate which genes are regulated by MtHDTs, RNA-seq analyses were conducted. We isolated RNA from nodules since it was not possible to collect sufficient primordium material, and especially because the majority of the MtHDTs RNAi primordia were blocked from developing, so the blocked primordia might have caused secondary effects. We collected apical part of nodules, including meristem and infection zone, as MtHDTs are preferentially expressed there. To dissect the apical part of nodules from the fixation zone, transgenic control and MtHDTs RNAi roots were inoculated with rhizobia expressing nifH::GFP. At the transition from the infection zone to the fixation zone, the nifH gene is switched on in the fixation zone (Gavrin et al., 2014), where MtHDTs are switched off. We will name the region containing meristem and infection zone the “nodule apex.”
Transcriptomes of control and MtHDTs RNAi nodule apices were analyzed and we detected the transcripts of ∼20,000 genes in each sample (Supplemental Data Set S1). The reduced expression levels of MtHDTs and MtPLT3,4 in MtHDTs RNAi nodule apices are consistent with reverse transcription quantitative polymerase chain reaction (RT-qPCR) data (Figure 3, A and C and Supplemental Data Set S1), indicating that RNA-seq data are reliable. To identify DEGs, we applied relatively stringent statistics and filtering (fold change > 4 and FDR P < 0.05). In total, 49 DEGs were identified between control and MtHDTs RNAi nodule apices (Supplemental Data Set S1).
To investigate whether HDTs control nodule development by regulating the same genes as in Arabidopsis roots, we first checked the expression of GA2ox genes, as they are targets of HDTs in Arabidopsis roots (Li et al., 2017). However, MtGA2ox genes, were not among the 49 DEGs (Supplemental Data Set S1), suggesting that HDTs regulate nodule and root development in a different way. To further test this, we compared the DEGs that are identified in Medicago nodule apices (n = 49) with those of Arabidopsis root tips (n = 217; Li et al., 2017). Gene orthology of the two species is well studied (van Velzen et al., 2018). About 63% (31 out of 49) of the Medicago DEGs have (an) putative orthologous gene(s) in Arabidopsis, but only the two HDT genes (MtHDT1/2, AtHDT1/2) were down-regulated in both RNAi experiments (Supplemental Data Set S2). This demonstrated that none of the DEGs that resulted from downregulation of HDTs were in common in Arabidopsis roots and Medicago nodules. We concluded that HDTs regulate nodule and root development in different ways.
To obtain insight into the biological functions of the identified 49 DEGs from nodule apices, we performed gene ontology (GO) analysis. This showed that genes encoding proteins with terpene synthase, methyltransferase, or oxidoreductase activities were among the enriched DEGs (Supplemental Figure S9).
MtHDTs possibly control nodule development by regulating MtHMGR1 expression
Two DEGs encode 3-hydroxy-3-methylglutaryl-coenzyme A reductases (MtHMGR1 and MtHMGR4). These two genes were downregulated 8.7 (MtHMGR1)- and 7.7 (MtHMGR4)-fold in MtHDTs RNAi nodule apices (Supplemental Data Set S1). Previously, it has been shown that knockdown of MtHMGR1 blocks nodule formation (Kevei et al., 2007). The function of MtHMGR1 in mature nodules has not been studied, but it has been shown to be an interactor of DOES NOT MAKE INFECTION 2 (MtDMI2; Kevei et al., 2007). Knockdown of MtDMI2 in nodules affects the intracellular colonization of rhizobia (Limpens et al., 2005), similar to that in MtHDTs RNAi nodules. Therefore, we focused on MtHMGR1.
To determine in which tissue MtHMGR1 is expressed and whether knockdown of MtHDTs affects its expression pattern, we performed RNA in situ hybridization on longitudinal sections of nodules harvested at 21 dpi. In control nodules, MtHMGR1 was expressed in nodule meristem and the infection zone, in the latter its expression only occurred in the infected cells (Figure 6, A). In MtHDTs RNAi nodules, MtHMGR1 had the same expression pattern (Figure 6, B), albeit at a markedly lower level (Supplemental Data Set S1).
Figure 6.
MtHDTs regulate the expression of MtHMGR1. A and B, In situ hybridization pattern of MtHMGR1 mRNA in ENOD12-EV (A) and MtHDTs RNAi (B) nodules. Arrowheads indicate infected cells in the infection zone. C and D, In situ hybridization pattern of MtHMGR1 mRNA in ENOD12-EV (C) and MtHDTs RNAi (D) nodule primordia. Arrows indicate noninfected cells. E, In situ hybridization pattern of MtHDT2 mRNA in wild-type nodule primordium. The arrow indicates a noninfected cell. F, RT-qPCR analysis of MtHMGR1 expression in ENOD12-EV control and MtHDTs RNAi nodule primordia. Data shown are mean ± SEM determined from three independent experiments. Nodules and nodule primordia were harvested at 21 and 5 dpi, respectively. In (A)–(E) longitudinal sections of nodules (A and B) or nodule primordia (C to E) were shown. Red dots are hybridization signals. Scale bars = 100 µm.
It has been shown that knockdown of MtHMGR1 blocks nodule primordium development, similar to the phenotype of the inoculated MtHDTs RNAi roots. We then asked whether the expression pattern and level of MtHMGR1 in nodule primordia was affected by knockingdown MtHDTs. To answer this, RNA in situ hybridization with MtHMGR1 probe set was performed on longitudinal sections of 5 dpi nodule primordia. In control nodule primordia (stage V), MtHMGR1 transcripts were very abundant in (future) meristems and infected cells (Figure 6, C). Expression pattern of MtHMGR1 in MtHDTs RNAi nodule primordia (stage V) resembled that of the control (Figure 6, D), albeit at a much lower level. RT-qPCR showed that the expression level of MtHMGR1 was 78% reduced (Figure 6, F). This is in line with the observation in mature nodules where knockdown of MtHDTs does not affect MtHMGR1 expression pattern, but only reduced its expression level (Figure 6, A and B and Supplemental Data Set S1).
In nodules, the expression pattern of MtHMGR1 coincides with that of the MtHDTs (Figures 2, 6, A and B). To test in nodule primordia whether MtHMGR1 and MtHDTs were expressed in the same cells, we performed RNA in situ hybridization with MtHDT2 probe set on longitudinal sections of 5 dpi nodule primordia. This revealed that in nodule primordia (stage V), MtHDT2 was also expressed in the future nodule meristem and infected cells (Figure 6, E), similar to MtHMGR1.
Taken together, our data showed that MtHDTs and MtHMGR1 were co-expressed during nodule development. Knockdown of MtHDTs did not affect the expression pattern of MtHMGR1, but only its expression level.
Discussion
In this study, we showed that the MtHDTs play a key role in both nodule primordium formation and nodule development. Knockdown of MtHDTs prevented primordium development and in nodules, it reduced meristem size and rhizobial colonization of cells. In both cases, these chromatin remodeling factors positively regulate the expression of MtHMGR1, which previously has been shown to be essential for nodule primordium formation (Kevei et al., 2007). The similar nodule primordium phenotype in MtHDTs and MtHMGR1 knockdown indicates that the decreased expression of MtHMGR1 is sufficient to explain the arrested nodule primordium development in MtHDTs RNAi. The mechanism by which MtHDT and MtHMGR1 control nodule (primordium) development is different from that involved in root development.
Knockdown of HDTs resulted in a similar root phenotype in Medicago and Arabidopsis (Supplemental Figure S4). This is in line with the fact that the MtHDTs and AtHDT1 and 2 have the same expression pattern in roots, and further, that pMtHDT2::GFP::HDT2 is sufficient to restore root development in an Arabidopsis hdt1hdt1hdt2hdt2 background. AtHDT1, 2 regulate root meristem size by repressing AtGA2ox2 (Li et al., 2017). Therefore, it is very probable that MtHDT2 has a similar function when expressed in Arabidopsis and it is likely that MtHDTs control Medicago root growth in a similar manner. If this is indeed the case, the mechanism by which MtHDTs regulate nodule meristem size is different, as expression of MtGA2oxs is not affected in MtHDTs RNAi nodule apices. Further, none of the Arabidopsis orthologs of the Medicago nodule DEGs is affected in the Arabidopsis HDTs RNAi roots. In addition, the expression pattern of the MtHDTs in nodules and roots is not similar. In nodules, the MtHDTs are expressed at equal levels in meristem and infection zone. The latter is equivalent to the root elongation zone. However, in roots, the MtHDTs are expressed at the highest level in the root meristem, whereas in the elongation zone, their expression level is very low.
It has been shown that the nodule and root developmental programs share transcription factors like PLETHORA and LBD16. It is possible that during the development of these two organs, different genes are regulated by these transcription factors. For example, during nodule development LBD16 interacts with a CCAAT box-binding protein Nuclear Factor-Y (NF-YA1), the latter is a nodule-specific transcription factor (Combier et al., 2006). The expression of LBD16 is directly regulated by NODULE INCEPTION (NIN; Schiessl et al., 2019; Soyano et al., 2019). NIN is a nodule-specific transcription factor as well (Marsh et al., 2007), indicating that the expression of LBD16 is also regulated differently during the development of both organs. Further, 96% of the transcriptional changes are shared between nin and lbd16 loss-of-function mutants. It is probable that the genes regulated by LBD16 during the development of both organs are not completely identical. Our study shows that chromatin remodeling factors HDTs are involved in root and nodule development, and their targets in these two processes are also different. So although root and nodule development share several regulators, it is possible that they have different functions.
Another chromatin remodeling factor, DNA demethylase (MtDME) is expressed in nodule infected cells. Knockdown of this gene does not decrease nodule number, but reduces the endoreduplication level of infected cells (Satge et al., 2016). MtDME is expressed at a low level in roots and its role in root development has not been studied, so whether it has a similar function in roots and nodules is unknown. During nodule development, MtDME first becomes active when rhizobial infection of cortical cells has already taken place. We show that MtHDTs are induced much earlier than MtDME, since the expression of MtHDTs is detected in nodule primordium cells prior to division (Figure 5 and Supplemental Figure S8). Similar to this, during initiation of lateral root primordium, AtHDT1/2 are induced in founder cells before the first cell division occurs (De Smet et al., 2008), suggesting that HDTs control the organogenesis of the two lateral organ primordia from the start.
It is well possible that more chromatin remodeling factors are shared between root and nodule development. Besides for HDTs, another five chromatin remodeling genes are upregulated in Arabidopsis early lateral root primordium, and their orthologs are upregulated in Medicago roots inoculated with rhizobia (Supplemental Table S2; Benedito et al., 2008; De Smet et al., 2008), so it will be worthwhile to compare their function during root and nodule development.
Knockdown of the three MtHDTs resulted in a nodule phenotype, whereas the only available Medicago hdt2 single mutant makes WT-like nodules, suggesting a functional redundancy of MtHDTs. Similarly, in Arabidopsis, both AtHDT1 and 2 control root development and leaf polarity (Ueno et al., 2007; Li et al., 2017). The redundancy might be due to the fact that AtHDTs as well as MtHDTs are the result of a recent gene duplication. In some monocots, such duplication has not occurred (Pandey et al., 2002; Grandperret et al., 2014), and knockdown of a single OsHDT701 (Figure 1, A) gene in rice enhances resistance to pathogens (Ding et al., 2012).
Silencing of MtHDTs resulted in a block of nodule primordium formation. We used the ENOD12 promoter to silence the MtHDTs. During nodule primordium initiation, the activation of this promoter could only be detected in pericycle and inner cortex once cell division had already occurred (Supplemental Figure S10). This implies that most likely, silencing is first effective when primordium formation has already been initiated. Therefore, it is certainly possible that MtHDTs are essential from the start of primordium initiation. In Arabidopsis roots, silencing of AtHDT1, 2 does not affect progression through the cell cycle. However, in most nodule primordia present in MtHDTs RNAi roots, DNA synthesis is blocked or markedly reduced, indicating that cell division is (being) blocked in these primordia (Figure 4). This further supports that HDTs have different roles in root and nodule development.
Although the MtHDTs are important for primordium development, still a few nodules formed on MtHDTs RNAi roots. Most likely, in these cases, expression of MtHDTs is not sufficiently reduced to block primordium development. In mature Medicago nodules, the approximately eight proximal cell layers with infected cells are derived from the primordium and not from the meristem (Xiao et al., 2014). In the few nodules formed on MtHDTs RNAi roots, rhizobial colonization is not affected in these infected cells derived from the primordium, but it is strongly reduced in cells of the infection zone derived from the nodule meristem. This difference in efficiency of colonization is in agreement with the idea that rhizobial infection in nodule cells is more stringently controlled than in primordium cells (Combier et al., 2006; Laporte et al., 2014).
The expression of MtHDTs in nodule meristem and infection zone is consistent with their function in colonization of infected cells, as well as in specifying nodule meristem properties. Considering that the MtHDTs RNAi nodule meristem is smaller, the reduced colonization in cells derived from this meristem might be the indirect effect of altered properties of the meristem cells. The cells of the meristem of these nodules still divide, whereas MtHDTs RNAi results in a prevention of cell division in primordia. However, the nodules are formed from primordia in which cell division is not (fully) blocked, most likely due to less reduction of the MtHDTs mRNA levels.
At the transition from the infection to fixation zone, MtHDT2 expression level dropped dramatically (Figure 2, A). At this transition, several other sudden changes occur, including accumulation of starch in the infected cells, collapse of the vacuole of the infected cells, and the induction of nifH genes of the rhizobia (Gavrin et al., 2014). So the sudden decrease of MtHDT transcripts and proteins supports the existence of a molecular switch at this transition.
MtHMGR1 is one of 49 DEGs, but its knockdown phenocopied MtHDTs RNAi, suggesting that the decreased expression of MtHMGR1 is sufficient to explain the MtHDTs RNAi nodule primordium phenotype. However, this does not exclude the possibility that other DEGs can also play an important role in nodule development. The decreased expression of MtHMGR1 is likely an indirect effect of MtHDTs knockdown, given that the activity of MtHDTs leads to condensed chromatin, which represses gene expression. We hypothesize that knockdown of MtHDTs might induce a transcriptional repressor of MtHMGR1, or alternatively, another chromatin remodeling factor that confers to the MtHMGR1 region a more condensed chromatin configuration. MtHMGR1 and its four paralogs are tandem repeats and they are all downregulated in MtHDTs RNAi (Supplemental Data Set S1). These clustered genes might have similar chromatin configurations, facilitating their regulation by chromatin remodeling factors.
Expression patterns of MtHMGR1 and MtHDTs overlapped in both nodule primordia and nodules (Figures 2, 6), indicating that MtHDTs regulate MtHMGR1 expression in a cell autonomous manner. MtHMGR1 is down-regulated in MtHDTs RNAi primordia as well as nodules, and the transcriptome studies show that all other MtHMGR members are downregulated in MtHDTs RNAi nodule apices (Supplemental Data Set S1). MtHMGRs encode enzymes that catalyze the rate-limiting step in the mevalonate pathway. This pathway leads to the synthesis of sterols and isoprenoids that give rise to several plant hormones, for example cytokinin, gibberellin, and abscisic acid (Chappell et al., 1995). Whether the disturbed isoprenoid biosynthesis results in the MtHDTs RNAi phenotype cannot be excluded. However, it has also been shown that MtHMGR1 knockdown affects Nod factor signaling, as it blocks rhizobium-induced Ca2+ spiking in the epidermis (Venkateshwaran et al., 2015). As Nod factor signaling is required for nodule primordium formation, this function can explain the primordium phenotype in both MtHMGR1 and MtHDTs RNAi (since in the latter the expression of MtHMGR1 is reduced). Nod factor signaling also occurs in the distal part of the infection zone. Knockdown of Nod factor receptor genes, as well as of an essential component of the Nod factor signaling cascade DMI2, results in reduced colonization of rhizobia in nodule cells (Limpens et al., 2005; Moling et al., 2014). This phenotype is similar to that of the MtHDTs RNAi nodules. So, in the case that MtHMGR1 is required for Nod factor signaling at early stages as well as in the nodule, its reduced expression can explain the MtHDTs RNAi nodule primordium and nodule phenotypes.
Materials and methods
Plant growth analysis, transformation, and rhizobial inoculation
Medicago ecotype Jemalong A17 and ENOD11::GUS stable line (Journet et al., 2001) were used in this study. Agrobacterium rhizogenes MSU440 mediated hairy root transformation was performed according to Limpens et al. (2004). The composite plants with transgenic roots were grown either in perlite saturated with low nitrate containing Farhaeus medium (Fahraeus, 1957), or on plates with agarose-based BNM medium (Ehrhardt et al., 1992), at 21°C in a 16 h:8 h, light:dark regime. Sinorhizobium meliloti 2011 or S. meliloti expressing nifH:GFP (Gavrin et al., 2014) liquid cultures were treated with 10 µM luteolin for 24 h, and then used to inoculate Medicago roots. Mature nodules were harvested at 21 dpi from roots of Medicago plants growing in perlite. Nodule primordia were harvested from spot inoculated roots of Medicago plants growing on plates at 2 or 5 dpi. Statistical analyses were done using SPSS software (the USA).
Phylogenetic tree construction
Gene accession numbers of HDTs are shown in Supplemental Table S1. For phylogenetic reconstruction, protein sequences were first aligned using MUSCLE (Edgar, 2004) implemented in Geneious Prime 2019 (New Zealand) using default parameters. After manual inspection, geneious tree builder was applied to generate the phylogeny by using neighbor-joining methods (Saitou and Nei, 1987).
Constructs
N-terminal fusions of MtHDTs with GFP under the control of their own promoters were constructed using MultiSite Gateway Technology (Thermo Fisher Scientific). The coding sequence (CDS) and putative promoter of each MtHDT were first PCR amplified by using Phusion high-fidelity DNA polymerase (Finnzymes), and nodule cDNA and genomic DNA were used as templates. The obtained PCR fragments were introduced into a pENTR-D-TOPO vector (Invitrogen). Each of the MtHDT promoters was cut out of the pENTR-D-TOPO vector using the NotI and AscI restriction enzymes, and then ligated with a BsaI digested pENTR4-1 vector (Invitrogen) containing GFP by using T4 DNA ligase (Thermo Fisher Scientific). The final pENTR4-1 vector with the MtHDT promoter and GFP, the corresponding pENTR-D-TOPO MtHDT CDS vector, and a pENTR2-3 vector containing a CaMV 35S terminator were recombined into the binary destination vector pKGW-RR-MGW, thereby creating pMtHDT::GFP::MtHDT constructs.
To create MtHDTs RNAi constructs, the PCR fragments of about 400–500 bp for each MtHDT CDS were amplified and then combined by subsequential PCR steps using primers with a complementary 15 bp overhang to generate one amplicon of all three MtHDTs fragments. The final product was introduced into a pENTR-D-TOPO vector (Invitrogen) and recombined in an inverted repeat orientation into the Gateway compatible binary vector pK7GWIWG2(II) driven by nodule specific ENOD12 promoter or CaMV 35S promoter (Limpens et al., 2005). The control vector [(ENOD12::Empty Vector (ENOD12-EV)] contained no coding DNA sequence. All primers used for cloning were listed in Supplemental Tables S3.1, S3.2.
Gene expression and RNA-Seq
Total RNA from transgenic nodules or nodule primordia was isolated using the plant RNA Easy Kit (Qiagen). cDNA was synthesized from 1 µg of isolated RNA by reverse transcription with random hexamer primers using the iScript Select cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative real-time PCR was performed in a 10-µL reaction system with SYBR Green super-mix (Bio-Rad). Ubiquitin was used as a reference gene. Primers used for quantitative real-time PCR are listed in Supplemental Table S3.3.
For RNA-Seq analyses, nodule meristem and infection zone were distinguished from the fixation zone under a fluorescent stereomacroscope (Leica) and manually dissected. Three independent experiments were conducted. Total RNA was extracted as described above. RNA was sequenced at BGI Tech Solutions (Hong Kong) using Hiseq2000 instrument. Sequencing data were uploaded to SRA database with BioProject No. PRJNA693426 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA693426), and were analyzed by mapping to the Medicago genome using CLC Genomics Workbench (Denmark). Gene expression levels were determined by calculating the RPKM (Reads Per Kilobase per Million mapped reads). DEGs are defined based on relatively stringent statistics and filtering (fold change > 4, FDR P < 0.05) within the CLC. GO enrichment analyses were performed using agriGO v2.0 (Tian et al., 2017).
RNA in situ hybridization
The nodules and nodule primordia were fixed with 4% (w/v) paraformaldehyde mixed with 5% (v/v) glutaraldehyde in 50-mM phosphate buffer (pH 7.4) and embedded in paraffin (Paraplast X-tra, McCormick Scientific). Sections of 7 μm were cut by RJ2035 microtome (Leica). RNA in situ hybridization was performed using Invitrogen ViewRNA ISH Tissue 1-Plex Assay kit (Thermo Fisher Scientific) according to the manual protocol (https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0018633_viewRNA_ISH_UG.pdf&title=VXNlciBHdWlkZTogVmlld1JOQSBJU0ggVGlzc3VlIEFzc2F5). RNA ISH probe sets were designed and produced by Thermo Fisher Scientific. Catalogue numbers of probe sets are the following: MtHDT1 is VF1-14234, MtHDT2 is VF1-18132, MtHDT3 is VF1-6000218, and MtHMGR1 is VF1-20373. Any probe set was omitted for a negative control. Slides were analyzed with an AU5500B microscope equipped with a DFC425c camera (Leica).
EdU staining
The composite plants with ENOD12-EV or MtHDTs RNAi transgenic roots were grown on BNM plates and spot inoculated with S. meliloti 2011 as described above. After 2 d, the inoculated root segments (∼0.3 cm) were submerged in liquid BNM medium with extra 1-g·L-1d-glucose, and were co-incubated with 10-µM EdU stock for 2 h on a shaker. The following washing and staining procedures were conducted according to Kotogany et al. (2010).
Microscopy and imaging
Root fragments and nodules were fixed as mentioned above. After that, they were washed with 0.1 M phosphate buffer three times for 15 min each, once with water for 15 min, and dehydrated for 10 min in 10%, 30%, 50%, 70%, 90%, and 100% (v/v) ethanol, and sequentially embedded in plastic Technovit 7100 (Heraeus Kulzer). Sections of 5 μm were made using a microtome (RJ2035, Leica), stained with 0.05% Toluidine Blue (Sigma), mounted in Euparal (Carl Roth), and analyzed with a Leica AU5500B microscope equipped with a DFC425c camera (Leica). Transgenic pMtHDT::GFP::MtHDT nodules and root segments were sectioned into 60-µm slices by vibratome (VT1000, Leica) and stained with propidium iodide for 2 min. Sections were then washed three times in phosphate buffer containing triton x-100 and mounted on slides with MQ water. All confocal images were acquired using Leica SP8 confocal laser scanning microscope (Leica, Germany). GFP and EdU signals were detected with excitation wavelength and detection windows of 488 and 500–530 nm, propidium iodide was detected with excitation wavelength and detection windows of 543 and 580–640 nm.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Table S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. MtHDT2 and MtHDT1 proteins are mainly localized in the nucleolus.
Supplemental Figure S2. MtHDT3 is expressed in root tips.
Supplemental Figure S3. Analysis of Mthdt Tnt1 mutants.
Supplemental Figure S4. Knockdown of MtHDTs reduces root meristem size in Medicago.
Supplemental Figure S5. Localization of MtHDT2 resembles that of AtHDT1, 2 in Arabidopsis root tips.
Supplemental Figure S6. Knockdown of MtHDTs reduces rhizobial colonization in the infection zone.
Supplemental Figure S7. Expression patterns of ENOD11::GUS in nodule primordia and lateral root primordia are different.
Supplemental Figure S8. MtHDT1 and MtHDT3 are expressed in nodules and nodule primordia.
Supplemental Figure S9. GO enrichment analysis of DEGs in MtHDTs RNAi nodule meristem and infection zone.
Supplemental Figure S10. Expression pattern of ENOD12::GUS during nodule primordium development.
Supplemental Table S1. Gene accessions used in the phylogenetic analysis.
Supplemental Table S2. The upregulated expression of chromatin remodeling genes in Arabidopsis lateral root primordia and Medicago nodule primordia.
Supplemental Table S3. Primers used in this study.
Supplemental Data Set S1. Gene expression map in the ENOD12-EV and MtHDTs RNAi nodule meristem and infection zone.
Supplemental Data Set S2. HDTs are the only overlapped DEGs in Medicago nodules and Arabidopsis roots.
Supplementary Material
Acknowledgments
The authors thank Defeng Shen from Max Plank Institute (Cologne, Germany) for the phylogenetic construction. They would also like to thank Xueyuan Leng and Nathalie Veltmaat who contributed as master students to this research.
Funding
This work was supported by grants from the European Research Council (2011-AdG-294790); the supporting plan for cultivating high level teachers in colleges and universities in Beijing (CIT&TCD20180317); the National Key Research and Development Program of China (2018YFD1000605); and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032).
Conflict of interest statement. None declared.
T.B. and H.L. designed the research and wrote the manuscript. H.L., S.S., Q.C., and O.K. performed the research and analyzed the data. Q.C. and O.K. revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ton Bisseling (ton.bisseling@wur.nl).
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55:504–513 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18:1269–1276 [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme-a reductase a rate-limiting step for isoprenoid biosynthesis in plants. Plant Physiol 109:1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20:3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322:594–597 [DOI] [PubMed] [Google Scholar]
- Ding B, Bellizzi MD, Ning YS, Meyers BC, Wang GL (2012) HDT701, a Histone H4 deacetylase, negatively regulates plant innate immunity by modulating Histone H4 acetylation of defense-related genes in rice. Plant Cell 24:3783–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214:30–36 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR (1992) Depolarization of alfalfa root hair membrane-potential by rhizobium-meliloti nod factors. Science 256:998–1000 [DOI] [PubMed] [Google Scholar]
- Fahraeus G (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16:374. [DOI] [PubMed] [Google Scholar]
- Franssen HJ, Vijn I, Yang WC, Bisseling T (1992) Developmental aspects of the Rhizobium-legume symbiosis. Plant Mol Biol 19:89–107 [DOI] [PubMed] [Google Scholar]
- Franssen HJ, Xiao TT, Kulikova O, Wan X, Bisseling T, Scheres B, Heidstra R (2015) Root developmental programs shape the Medicago truncatula nodule meristem. Development 142:2941. [DOI] [PubMed] [Google Scholar]
- Gavrin A, Kaiser BN, Geiger D, Tyerman SD, Wen ZY, Bisseling T, Fedorova EE (2014) Adjustment of host cells for accommodation of symbiotic bacteria: vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell 26:3809–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139:883–893 [DOI] [PubMed] [Google Scholar]
- Grandperret V, Nicolas-Frances V, Wendehenne D, Bourque S (2014) Type-II histone deacetylases: elusive plant nuclear signal transducers. Plant Cell Environ 37:1259–1269 [DOI] [PubMed] [Google Scholar]
- Han ZF, Yu HM, Zhao Z, Hunter D, Luo XJ, Duan J, Tian LN (2016) AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Pineiro M, Cubas P, Martinez-Zapater JM (2009) Chromatin remodeling in plant development. Int J Dev Biol 53:1581–1596 [DOI] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14:737–748 [DOI] [PubMed] [Google Scholar]
- Kevei Z, Lougnon G, Mergaert P, Horvath GV, Kereszt A, Jayaraman D, Zaman N, Marcel F, Regulski K, Kiss GB, et al. (2007) 3-hydroxy-3-methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19:3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotogany E, Dudits D, Horvath GV, Ayaydin F (2010) A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, et al. (2014) The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot 65:481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HC, Torres-Garcia J, Latrasse D, Benhamed M, Schilderink S, Zhou WK, Kulikova O, Hirt H, Bisseling T (2017) Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number. Plant Cell 29:2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci U S A 102:10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992 [DOI] [PubMed] [Google Scholar]
- Lusser A, Brosch G, Loidl A, Haas H, Loidl P (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277:88–91 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144:324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moling S, Pietraszewska-Bogiel A, Postma M, Fedorova E, Hink MA, Limpens E, Gadella TWJ, Bisseling T (2014) Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26:4188–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30:5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrere S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77:817–837 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Samac DA, Tesfaye M, Dornbusch M, Saruul P, Temple SJ (2004) A comparison of constitutive promoters for expression of transgenes in alfalfa (Medicago sativa). Transgenic Res 13:349–361 [DOI] [PubMed] [Google Scholar]
- Satge C, Moreau S, Sallet E, Lefort G, Auriac MC, Rembliere C, Cottret L, Gallardo K, Noirot C, Jardinaud MF, et al. (2016) Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nature Plants 2 [DOI] [PubMed] [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD, et al. (2019) NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr Biol 29:3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Kawaguchi M, Hayashi M (2019) A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 366:1021. [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan HY, You Q, Yi X, Du Z, Xu WY, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45:W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Poole PS (2013) Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805 [DOI] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19:445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378:62–65 [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu FJ, Rutten L, van Zeijl A, Liu W, Santuari L, Cao QQ, Sharma T, Shen DF, et al. (2018) Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc Natl Acad Sci U S A 115:E4700–E4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci U S A 106:11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M, Jayaraman D, Chabaud M, Genre A, Balloon AJ, Maeda J, Forshey K, den Os D, Kwiecien NW, Coon JJ, et al. (2015) A role for the mevalonate pathway in early plant symbiotic signaling. Proc Natl Acad Sci U S A 112:9781–9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, Tarayre S, Roudier F, Mergaert P, Kondorosi A, et al. (2003) Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15:2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141:3517–3528 [DOI] [PubMed] [Google Scholar]
- Xiao TT, van Velzen R, Kulikova O, Franken C, Bisseling T (2019) Lateral root formation involving cell division in both pericycle, cortex and endodermis is a common and ancestral trait in seed plants. Development 146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.