Figure 4.
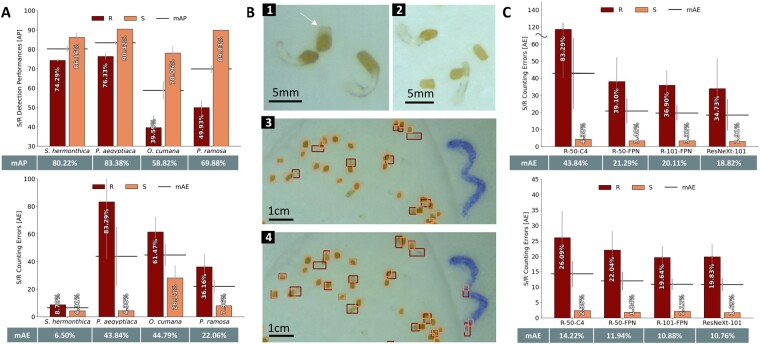
Generalization capabilities of the developed object detection algorithm for measuring germination of other parasitic seeds. A, Assessment of the performance of SeedQuant backbone (R-50-C4, developed on Striga seeds) on O. ramosa, P. cumana, and P. aegyptiaca seeds images (32 images for each seed type), in comparison with the hand annotations for the S/R (seed/radicle) annotation approach. Evaluation of (upper) the detection performances and (lower) the counting performances. The horizontal bars on the bar graphs represent the mAE/mAP, respectively, for each architecture and its corresponding value is reported in the table below. The error bars that indicate standard error. B, (1) Visualization of the P. aegyptiaca seeds morphology (nongerminated indicated by an arrow), showing their extra morphological structure, in comparison with (2) Striga seeds. Visualization of the bounding boxes placed by hand (3) and by the R-40-C4 algorithm (4) (seeds in light red, radicle in dark red). The direct processing of the P. aegyptiaca images by the Striga-based algorithm led to a lot of false positives in radicle detection, as the presence of the additional white part of the seed coat was classified as a very short radicle. C, Assessment of the counting performance of the different Faster-CNN backbone architectures (R-50-C4, R-50-FPN, R-101-FPN, ResNeXt-101) used for SeedQuant development on P. aegyptiaca seeds, based on the S/R object classification: (upper) prior fine-tuning on 32 images and (lower) after fine-tuning on 30 images, using 31 P. aegyptiaca seeds images for an additional training. The horizontal bars on the bar graphs represent the mean value of each architecture, its corresponding value is reported in the table below. The error bars that indicate standard error.