Abstract
Stomatal density (SD) and stomatal complex area (SCA) are important traits that regulate gas exchange and abiotic stress response in plants. Despite sorghum (Sorghum bicolor) adaptation to arid conditions, the genetic potential of stomata-related traits remains unexplored due to challenges in available phenotyping methods. Hence, identifying loci that control stomatal traits is fundamental to designing strategies to breed sorghum with optimized stomatal regulation. We implemented both classical and deep learning methods to characterize genetic diversity in 311 grain sorghum accessions for stomatal traits at two different field environments. Nearly 12,000 images collected from abaxial (Ab) and adaxial (Ad) leaf surfaces revealed substantial variation in stomatal traits. Our study demonstrated significant accuracy between manual and deep learning methods in predicting SD and SCA. In sorghum, SD was 32%–39% greater on the Ab versus the Ad surface, while SCA on the Ab surface was 2%–5% smaller than on the Ad surface. Genome-Wide Association Study identified 71 genetic loci (38 were environment-specific) with significant genotype to phenotype associations for stomatal traits. Putative causal genes underlying the phenotypic variation were identified. Accessions with similar SCA but carrying contrasting haplotypes for SD were tested for stomatal conductance and carbon assimilation under field conditions. Our findings provide a foundation for further studies on the genetic and molecular mechanisms controlling stomata patterning and regulation in sorghum. An integrated physiological, deep learning, and genomic approach allowed us to unravel the genetic control of natural variation in stomata traits in sorghum, which can be applied to other plants.
High-throughput phenotyping using deep learning tools integrated with genome-wide association studies revealed genes that control SD and area in grain sorghum.
Introduction
Stomata are microscopic pores on the leaf surface that facilitate gas exchange between the leaf and the atmosphere, most notably CO2 and water vapor. Stomata exert a substantial influence on crop productivity through photosynthesis and water use, which are driven by stomatal conductance (gs) (Field et al., 1983; Franks and Farquhar, 2007; Franks and Beerling, 2009; Dow et al., 2014a, 2014b; Medeiros et al., 2016). Considering the influence of stomatal characteristics (shape, density, and area, distribution of stomata [between Ab and Ad]) on crop productivity under a range of environmental conditions (Fanourakis et al., 2015; Faralli et al., 2019), genetic variation in stomatal traits is considered a key target for crop improvement (Shimazaki et al., 2007; Kim et al., 2010). Stomatal response varies from short-term (opening and closing of stomata driven by guard cell expansion and shrinkage; Shimazaki et al., 2007) to long-term morphological changes in stomatal density (SD) or area due to environmental (Hetherington and Woodward, 2003; Buckley et al., 2020) and internal signals (Chater et al., 2011; Kinoshita et al., 2011; Chater et al., 2017). A substantial variation in the SD between the Ad (upper) and Ab (lower) surfaces exists in crops (Bertolino et al., 2019), including sorghum (Liang et al., 1975).
SD and stomatal size/complex area are two key parameters studied in both model and non-model plant species (Doheny-Adams et al., 2012; Hepworth et al., 2018; Bertolino et al., 2019; Buckley et al., 2020). A positive relationship between SD and gs, has been reported both within (Reich, 1984; Muchow and Sinclair, 1989; Tanaka et al., 2010; Carlson et al., 2016) and between species (Anderson and Briske, 1990; Pearce et al., 2006). Furthermore, a negative relationship between SCA/size and SD has been observed both in C3 and C4 species (Kawamitsu et al., 1996). This suggests that SD is not the only parameter regulating the balance between water loss and carbon uptake. SCA/size is defined as a product of guard cell length and width (Franks and Beerling, 2009; Drake et al., 2013). The width of the guard cells is reported to be more dynamic, compared to the stomata length, in responding to changes in the environmental conditions during the day (Lawson et al., 1998; Lawson and Blatt, 2014).
The genetic manipulation of pathways associated with stomatal development, patterning, and regulation is demonstrated in the model dicot Arabidopsis thaliana (Chater et al., 2011; Hepworth et al., 2018) and crops (Faralli et al., 2019), to optimize water use and yield (Buckley et al., 2020). The possibility of improving water-use efficiency (WUE) by reducing SD on the leaf surface is demonstrated in dicots (Yu et al., 2008; Yoo et al., 2010; Hepworth et al., 2015) and monocots (Hughes et al., 2017; Caine et al., 2019). Reduced SD in major food crops such as rice, (Oryza sativa L.; Caine et al., 2019), wheat (Triticum aestivum;Dunn et al., 2019), and barley (Hordeum vulgare;Hughes et al., 2017) resulted in increased WUE and drought tolerance through reduction in water loss, without affecting yield. Alternatively, theoretical and experimental evidence suggest that larger stomata are generally not effective for rapid gas exchange compared with smaller stomata, due to the greater pore depth (Raven, 2014; Faralli et al., 2019). A negative relationship between the stomata area and WUE is shown in Arabidopsis (Dittberner et al., 2018). This is because larger stomata take a longer time to close compared to smaller stomata, leading to additional water loss, which increases the amount of water expended per unit of biomass produced (Faralli et al., 2019). Taken together, a combination of smaller stomata with high density would translate to higher gs and productivity under nonstress conditions (Franks and Beerling, 2009; Henry et al., 2019), while an optimized number and area of stomata would be beneficial under water-limited conditions (Lawson and Blatt, 2014; Leakey et al., 2019). Thus, exploring the phenotypic diversity and genetic basis of stomatal traits would provide useful information to improve productivity and stress tolerance in sorghum (Sorghum bicolor).
Stomatal characteristics, including SD and SCA have been studied using manual low throughput methods in crops exposed to different environments (Gitz and Baker, 2009). However, genetic architecture controlling stomatal traits and their responses to different environments is not known in sorghum. In addition, the diversity in stomatal traits is largely unexplored or utilized in breeding programs due to a cumbersome phenotyping protocol, which requires substantial investment of resources. For example, manual phenotyping of stomatal count involves obtaining stomatal imprints, imaging of the specimen, and manual counting of stomatal numbers, with the latter requiring most time and effort (Fetter et al., 2019; Sakoda et al., 2019). In the current genomic era, phenotyping of traits has been identified as a substantial bottleneck compared to generating large genome sequence datasets (Hudson, 2008). Recently, several computer vision-based automated phenotyping tools have been developed to overcome this challenge by automated detection of stomata, including Cascade object detection algorithm (Higaki et al., 2014; Laga et al., 2014; Duarte et al., 2017; Jayakody et al., 2017), AlexNet-based deep convolutional neural network (Fetter et al., 2019) and You Only Look Once (Casado and Heras, 2018). Several approaches and tools for quantifying stomatal variations based on images have been proposed (Dittberner et al., 2018; Fetter et al., 2019; Sakoda et al., 2019). However, previous methods have followed the object detection approach instead of the more precise semantic object segmentation (see “Materials and methods”). To address this limitation, we trained the Mask Region-based Convolutional Neural Network (Mask R-CNN) algorithm to automatically predict labels for future images, to segment the stomata in an image to identify and count stomata, and to determine the SCA.
Sorghum is generally grown in arid and semi-arid regions, and hence its productivity depends on timing and amount of rainfall. This poses a crucial challenge to sorghum grown in USA, Sub-Saharan Africa, India, and other regions in the world (Leff et al., 2004). Despite their adaptation to arid conditions, sorghum hybrids are shown to be susceptible to harsh environments during different stages of the crop growth (Tack et al., 2017). Given that C4 crops including sorghum have evolved and adapted to hot and arid conditions (Osborne and Freckleton, 2009), they provide an excellent opportunity to investigate natural variability in SD and area under field conditions. To date, there has not been an attempt to map the genetic loci associated with stomatal traits using the grain sorghum association panel (SAP). Thus, we hypothesized that integration of physiology, deep learning, and genomic approaches would help us understand the genetic architecture of stomatal traits in grain sorghum. The Genome-Wide Association Study (GWAS) is an efficient and powerful tool for unraveling the genetic basis of complex traits compared to bi-parental mapping in sorghum (Casa et al., 2008; Morris et al., 2013). Hence, a GWAS approach was used to identify genetic loci or favorable alleles (FAs) underlying SD and area in grain sorghum.
In this study, we characterized the genetic variation for stomatal traits using SAP in two environments in Kansas, USA. Additionally, we integrated the high-throughput deep learning tools and classical phenotyping methods to map genomic regions associated with stomatal number and area. Specific objectives were to (1) develop, test, and validate a fully automated deep learning tool for high-throughput phenotyping of Ab and Ad SD and SCA on a diversity panel; (2) comparative assessment of the stomatal traits obtained with deep learning (predicted) and manual methods; and (3) use GWAS results to determine the level of agreement between deep learning and manual methods. Lastly, the performance of accessions carrying contrasting haplotypes for SD on chromosome 6 was independently characterized, and differences in gas exchange were quantified, under field conditions.
Results
To investigate the natural variation in SD, SCA, and single leaf area (SinLA) in sorghum, a SAP representing a large variation in the geographical origin, races, and genetic diversity was used (Harlan and Wet, 1972; Casa et al., 2008; Morris et al., 2013). Stomatal traits are influenced by developmental stage, genetic background, and the environment (Liang et al., 1975). Considering these internal and external factors, we have characterized a common set of accessions (n = 311, Supplemental Table S1) in two environments (Env.) in Kansas (Env. 1—Manhattan and Env. 2—Hays; Supplemental Figure S1) in Exp. 1. Furthermore, the performance of accessions carrying contrasting haplotypes for SD on chromosome 6 was reconfirmed in the same environments in Exp. 2. A schematic overview of the study is visualized in Figure 1, A–C. Based on 6 years (2010–2016) of annual average difference in precipitation between environments, Env. 1 was considered as high rainfall environment (754 mm), and Env. 2 (449 mm) was considered as low rainfall environment (https://mesonet.k-state.edu/). All weather variables observed from planting to sampling dates at the experimental sites are given in Supplemental Figure S1, A–C (2017-Exp. 1) and Supplemental Figure S1D–F (2018-Exp. 2).
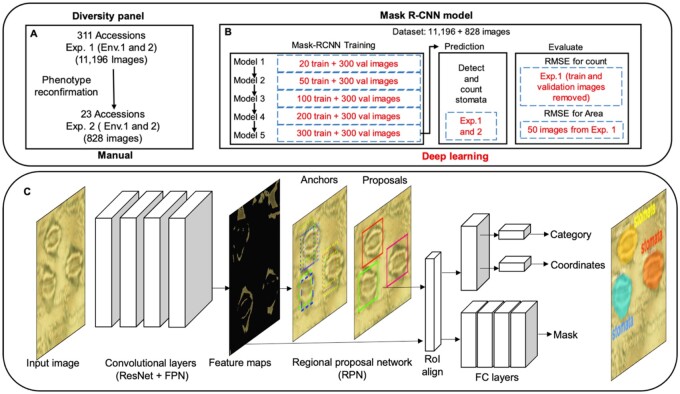
Figure 1.
Schematic overview of the study. A, Phenotyping of the SAP for SD and SCA in two environments (Env. 1—Manhattan and Env. 2—Hays) for two years (Exp. 1 in 2017 and Exp. 2 in 2018; see Supplemental Figure S1). B, Mask R-CNN models trained for predicting Ab and Ad stomatal number and complex area. Train and validate (val) images indicate the number of images used for training and validating the Mask R-CNN model trained. C, Mask R-CNN, a deep learning framework for stomata instance segmentation and stomata count. The network architecture contains convolutional layers (left) and fully connected layers (right), shown as rectangular cuboids in the figure. The size of each cuboid indicates the dimensionality of the corresponding layer. The connections between layers are represented through arrows. Detailed procedure followed to train, validate, and select the best model is provided in the Supplemental Figure S2.
Classical and deep learning methods in evaluating the SD and SCA
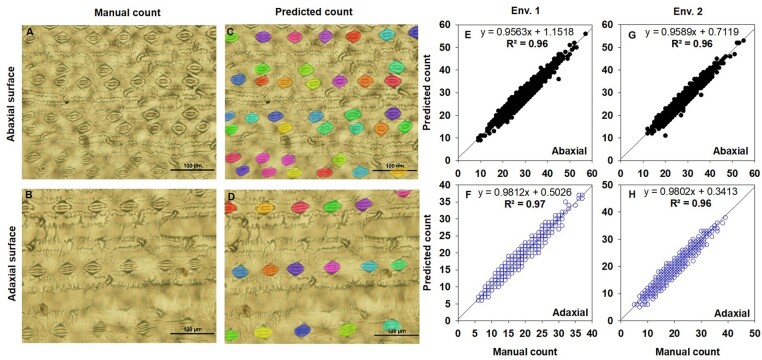
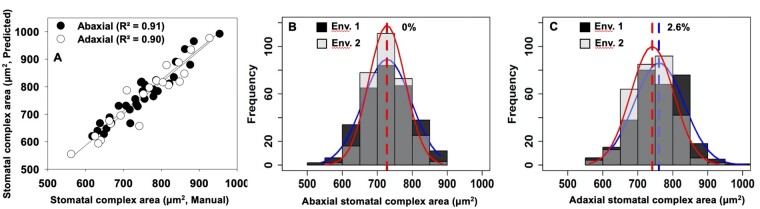
In addition to manual counting of stomata on 11,196 images, we developed a deep learning tool to extract SD and SCA automatically, using the Mask R-CNN model (Figure 1, see “Materials and methods”). The Mask R-CNN model was developed by experimenting with datasets of different sizes (Supplemental Figure S2) and identifies, classifies, and counts the number of stomata and measures SCA of all stomata in an image (Figure 1, B and C). The model trained with 300 images and validated with 300 additional images had the lowest validation loss (Supplemental Figure S2B). A strong correlation was observed between the human measured and predicted values of the remaining images in our dataset (r = 0.98 with Root Mean Square Error (RMSE)= 1.76; Supplemental Figure S2, C and D). This model also gave the lowest error (Supplemental Figure S2C), and was hence considered to explore the genetic diversity in the SD and SCA (Table 1). A comparison between manual (observed; Figure 2, A and B) and automated (prediction; Figure 2, C and D) stomata counts recorded a significant positive association between methods for Ab (R2 = 0.96 and R2 = 0.96; Figure 2, E and G) and Ad SD (R2 = 0.97 and R2 = 0.96; Figure 2, F and H) in Env. 1 and Env. 2, respectively. The broad-sense heritability (H2) values of the Ab (0.72) and Ad (0.72) SD were the same between methods (Table 1). There was a strong relationship between the predicted and human measured SCA for Ab (R2 = 0.91) and Ad (R2 = 0.90) leaf surfaces (Figure 3A). Based on this strong relationship between the manual and predicted values for Ab and Ad SCA, the predicted data on the entire diversity panel were considered for further analysis (Figure 3).
Table 1.
ANOVA and variation in phenotypic traits using classical phenotyping and deep learning methods in SAP in environments 1 and 2
| Trait | Acronym | G | E | G × E | Environment 1 |
Environment 2 |
H2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | h2 | Minimum | Maximum | h2 | ||||||
| Manual (Classical method) | |||||||||||
| Ab SD (mm−2) | SDAb | <0.001 | <0.001 | <0.001 | 53.7 | 180.56 | 0.44 | 64.81 | 174.07 | 0.43 | 0.72 |
| Ad SD (mm−2) | SDAd | <0.001 | <0.001 | <0.001 | 41.2 | 118.06 | 0.49 | 41.20 | 128.70 | 0.28 | 0.72 |
| Ab stomatal number (×106 per leaf) | SNAb_LA | <0.001 | <0.001 | <0.001 | 0.89 | 9.33 | 0.39 | 1.31 | 8.15 | 0.43 | 0.79 |
| Ad stomatal number (×106, per leaf) | SNAd_LA | <0.001 | 0.005 | <0.001 | 0.66 | 5.70 | 0.36 | 1.03 | 5.21 | 0.10 | 0.78 |
| Single leaf area (cm2) | SinLA | <0.001 | <0.001 | <0.001 | 72.64 | 813.79 | 0.38 | 117.88 | 673.70 | 0.28 | 0.84 |
| Predicted (deep learning method) | |||||||||||
| Ab SD (mm−2) | SDAb | <0.001 | <0.001 | <0.001 | 52.78 | 177.78 | 0.46 | 66.20 | 168.52 | 0.45 | 0.72 |
| Ad SD (mm−2) | SDAd | <0.001 | <0.001 | <0.001 | 41.67 | 118.98 | 0.52 | 38.89 | 125.46 | 0.28 | 0.72 |
| Ab stomatal number (×106 per leaf) | SNAb_LA | <0.001 | <0.001 | <0.001 | 0.90 | 9.36 | 0.38 | 1.30 | 7.89 | 0.43 | 0.79 |
| Ad stomatal number (×106 per leaf) | SNAd_LA | <0.001 | NS | <0.001 | 0.66 | 5.71 | 0.36 | 1.05 | 5.09 | 0.08 | 0.78 |
| Ab SCA (μm2) | SCAAb | <0.001 | NS | <0.001 | 538.01 | 879.11 | 0.45 | 550.16 | 885.21 | 0.33 | 0.74 |
| Ad SCA (μm2) | SCAAd | <0.001 | <0.001 | <0.001 | 574.77 | 1008.65 | 0.56 | 560.69 | 932.92 | 0.36 | 0.76 |
Probability values of the effects of genotype (G), environment (E), and their interaction (G × E) for all of the traits measured by ANOVA. NS indicates nonsignificant.
h2 indicates marker-based narrow sense heritability using Genome Association and Prediction Integrated Tool.
H 2 indicates the broad-sense heritability estimated considering the proportion of phenotypic variance that is due to genetic variance. Mask R-CNN, a framework of deep learning method was used to predict the SD and SCA.
Figure 2.
Results of SD (per image) following manual and deep learning methods. Comparison of ground-truth images (A and B) and deep learning segmentation results (C and D, predicted stomata highlighted in colors). Relationship of the SD obtained from manual count with predicted count obtained from the deep learning method (E and G—Ab; F and H—Ad). SAP was characterized in two environments (Env. 1 and Env. 2). A total of 11,196 (in Exp. 1) and 828 (Exp. 2) images were used to manually count stomata and generate the observational ground-truth SD data. The same sets of images were used to predict the SD with the deep learning method, as illustrated in Figure 1. A–D, bars = 100 µm.
Figure 3.
Relationship of observed SCA (μm2) with the corresponding data obtained using deep learning method (A). SCA was predicted using the deep learning approach on the entire SAP grown in Env. 1 and Env. 2 in 2017. Panels “B and C” show the distribution (Env. 1—blue line, dark gray bars; Env. 2—red line, light gray bars; intermediate gray bars indicate the overlap between the environments) of Ab and Ad stomata complex area, respectively. The vertical dotted lines on the histograms show population mean values in Env. 1 (blue) and Env. 2 (red). Values represent the positive percentage change in mean phenotypic value with respect to Env. 1 = [(mean trait value of Env. 1 − mean trait value of Env. 2)/mean trait value of Env. 1]×100.
Large natural variation for stomatal traits and SinLA in sorghum
The SAP showed significant variation among all the studied traits across environments (Table 1). Ab and Ad SD of manual (Supplemental Figure S3) and predicted (Supplemental Figure S4) counts were close to normal distributions across environments. Similarly, predicted SCA was also normally distributed in both environments (Figure 3, B and C). In general, SD was significantly higher by 1.5 times in the Ab surface than the Ad; inversely, SCA was smaller on the Ab surface than Ad in sorghum (Table 1).
To examine the relationship among traits and factors contributing to the total phenotype variation, 11 traits (5 manual and 6 predicted; Table 1) were used to perform principal component analyses (PCAs) within each environment (Supplemental Figure S5). In Env. 1, the first two principal components (PCs) cumulatively explained >79% variance (Supplemental Figure S5A). The PCA results of Env. 2 explained that 75% of the trait variance was caused by PC1 and PC2 (Supplemental Figure S5B). Similar to Env. 1, the loading value on PC1 was high for SinLA and derived traits (Ab and Ad stomatal number per leaf) and the PC2 for stomata density (mm−2; Supplemental Figure S5B). In both environments, the loading on PC2 was positive for SD (mm−2), and negative for loading on SCA (μm2). Similarly, there was a strong negative correlation of Ab SD with Ab SCA (Env. 1: r = −0.53, P < 0.001; Env. 2: r = −0.47, P < 0.001) in both environments (Supplemental Figure S6, A and B), whereas Ab SD or SCA exhibited significant (P < 0.01 to P < 0.001) positive correlations with Ad SD or SCA in both environments (Supplemental Figure S6).
Highlights of GWAS
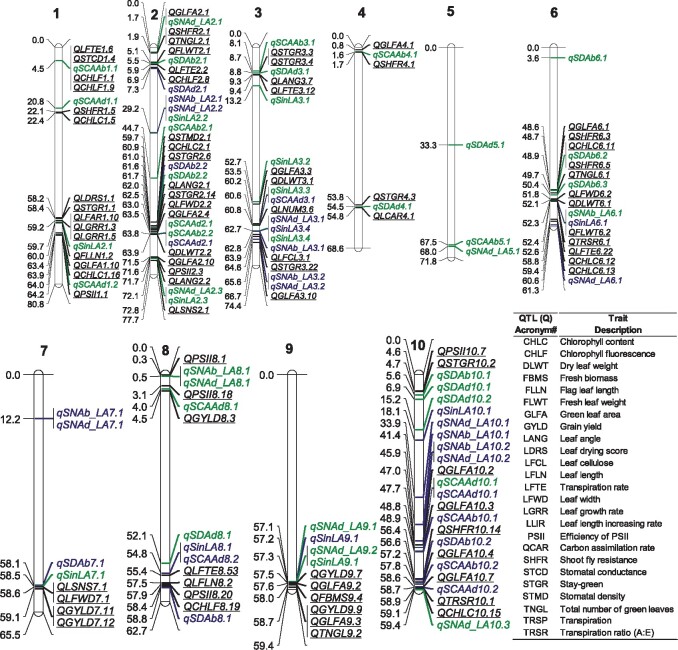
GWAS using a mixed linear model (MLM) provided substantial insights into the genetic architecture of stomatal traits in sorghum. Manhattan and quintile–quintile (Q–Q) plots of all traits from both manual and predicted datasets across environments are presented in Supplemental Figures S7–S10. A list of significant single nucleotide polymorphisms (SNPs) was detected and their ranking (based on P-values for the corresponding chromosome) in stomatal traits from both manual and predicted datasets is given in the Supplemental Table S2. Based on the reported mean distance of the linkage disequilibrium (LD) decay rates of up to 150 kb in sorghum (Morris et al., 2013; Ortiz et al., 2017; Moghimi et al., 2019), the SNP with the lowest P-value within 100 kb is considered as a candidate SNP (cSNP) to represent that locus (see “Materials and methods”). With this criterion, we identified 71 cSNPs (38 were environment specific and 33 shared between Env. 1 or Env. 2) for all the traits across environments (Figure 4; Supplemental Table S3). Half of the detected genetic loci (36/71) were overlapping with those previously reported for gas exchange and other related physiological traits (Figure 4; Supplemental Table S4).
Figure 4.
Summary of the associated genetic loci for all the investigated traits, as revealed by GWAS. The lines (black, green, and blue) in chromosomes denote the physical position (Mb) of the cSNPs that were identified in the study. The position (in Mb) of the locus is presented on the left side of each chromosome. The locus name is shown on the right: loci found in Env. 1 [green], Env. 2, [blue] in the current study and previously reported QTLs (underlined in black) were associated with similar or closely related traits, including gas exchange, leaf morphology, and yield traits. Previously reported genomic regions or QTL IDs given in the map were obtained from https://aussorgm.org.au/sorghum-qtl-atlas/ (Mace et al., 2019), see Supplemental Table S4. SDAb, abaxial stomatal density; SDAd, adaxial stomatal density; SNAb_LA, abaxial stomatal number per single leaf (×106); SNAd_LA, adaxial stomatal number per single leaf (×106); SCAAb, stomata complex area of abaxial; SCAAd, stomata complex area of adaxial; and SinLA, single leaf area. # QTL (Q) acronym for the previously reported traits.
GWAS findings between manual and predicted SD
Statistically significant correlations (r = 0.99, P < 0.001) were observed with Ab SD obtained using manual and predicted datasets in both environments (Supplemental Figure S6). Likewise, a comparison of GWAS results between manual and predicted methods for Ab SD resulted in identifying the same cSNPs (within an environment) on chromosomes 2, 6, 7, 8, and 10 (Table 2). Despite slight variation in the cSNP ranking and P-values (Supplemental Table S3), cSNPs on different chromosomes were common for Ab SD identified with manual and predicted methods in Env. 1 and Env. 2 (Table 2). For example, cSNP, S6_50424601 of qSDAb6.3 was commonly detected for Ab SD on chromosome 6 using both methods in Env. 1. cSNP, S10_56551896 of qSDAb10.2 was the most significant SNP identified (P = 6E-06) using predicted data, while the same SNP ranked second (P = 8E-06) with manual data in Env. 1 (Table 2). In Env. 2, cSNPs, S8_58766382 (qSDAb8.1) and S7_58134055 (qSDAb7.1) were the most significant SNPs identified for Ab SD using both manual and predicted datasets (Table 2). Similarly, S8_513583 and S3_65626261 are the most significant common cSNPs detected for Ab stomatal number per leaf in Env. 1 and 2, respectively, across both methods (Table 2). The effects of all cSNPs were almost similar across both methods. Additionally, the comparative GWAS analysis for the Ad surface showed the same genomic regions across phenotyping methods, with slight variations in the ranking of cSNP (Supplemental Figure S8; Supplemental Table S3). Narrow-sense heritability (h2) values of traits were similar in both datasets, for example, h2 of Ab SD (mm−2) was 0.44 and 0.46 (in Env. 1) and 0.43 and 0.45 (in Env. 2) for manual and predicted methods, respectively (Table 1).
Table 2.
Summary of genetic loci detected for Ab SD using manual and deep learning methods in Environments 1 and 2
| Locus | Environment | cSNP | Alleles | MAF | Manual |
Prediction |
FA | Genesa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | R2 | AE | Rank | P-value | R2 | AE | Rank | |||||||
| qSDAb6.3 | Env. 1b | S6_50424601 | T:C | 0.06 | 4E-06 | 0.14 | −13.01 | 1 | 9E-06 | 0.13 | −12.13 | 2 | T | 15 |
| qSDAb10.2 | Env. 1 | S10_56551896 | C:T | 0.28 | 8E-06 | 0.13 | −6.99 | 2 | 6E-06 | 0.14 | −6.87 | 1 | C | 17 |
| qSDAb10.1 | Env. 1 | S10_5625444 | A:C | 0.08 | 2E-05 | 0.13 | 10.45 | 3 | 1E-05 | 0.13 | 10.20 | 3 | C | 13 |
| qSDAb2.2 | Env. 1b | S2_61654537 | A:C | 0.07 | 3E-05 | 0.13 | −11.58 | 6 | 2E-05 | 0.13 | −11.34 | 5 | A | 11 |
| qSDAb2.1 | Env. 1 | S2_5471657 | G:A | 0.05 | 4E-05 | 0.12 | 12.76 | 8 | 2E-05 | 0.13 | 12.96 | 4 | A | 7 |
| qSDAb8.1 | Env. 2 | S8_58766382 | C:T | 0.33 | 7E-06 | 0.16 | −5.37 | 1 | 2E-06 | 0.18 | −5.59 | 1 | C | 13 |
| qSDAb7.1 | Env. 2b | S7_58134055 | T:C | 0.05 | 7E-06 | 0.16 | −11.07 | 2 | 4E-06 | 0.17 | −11.26 | 2 | T | 4 |
| qSDAb6.2 | Env. 2b | S6_48877403 | G:T | 0.07 | 2E-05 | 0.16 | −9.69 | 6 | 6E-05 | 0.16 | −9.05 | 7 | G | 18 |
| qSDAb2.2 | Env. 2b | S2_61589467 | A:G | 0.16 | 4E-05 | 0.15 | −6.76 | 7 | 5E-05 | 0.16 | −6.63 | 6 | A | 11 |
| qSDAb6.1 | Env. 2 | S6_3637502 | A:T | 0.06 | 6E-05 | 0.15 | −8.04 | 9 | 1E-04 | 0.15 | −7.71 | 10 | A | 6 |
| qSNAb_LA8.1 | Env. 1b | S8_513583 | C:G | 0.16 | 4E-06 | 0.27 | 0.59 | 1 | 4E-06 | 0.27 | 0.59 | 1 | G | 17 |
| qSNAb_LA6.1 | Env. 1 | S6_52304035 | G:T | 0.11 | 1E-05 | 0.26 | 0.57 | 6 | 8E-06 | 0.27 | 0.59 | 4 | T | 12 |
| qSNAb_LA2.1 | Env. 1 | S2_29172722 | C:G | 0.08 | 2E-05 | 0.26 | −0.66 | 8 | 2E-05 | 0.27 | −0.67 | 7 | C | 0 |
| qSNAb_LA10.1 | Env. 1 | S10_41443418 | A:G | 0.15 | 2E-05 | 0.26 | −0.55 | 11 | 4E-05 | 0.26 | −0.53 | 14 | A | 4 |
| qSNAb_LA3.1 | Env. 1 | S3_62821056 | C:G | 0.13 | 8E-05 | 0.25 | −0.54 | 18 | 5E-05 | 0.26 | −0.55 | 15 | C | 8 |
| qSNAb_LA3.2 | Env. 2b | S3_65626261 | A:G | 0.48 | 9E-06 | 0.26 | 0.30 | 1 | 2E-05 | 0.27 | 0.29 | 1 | G | 17 |
| qSNAb_LA10.2 | Env. 2 | S10_45929740 | A:C | 0.09 | 2E-05 | 0.26 | 0.50 | 2 | 2E-05 | 0.27 | 0.49 | 2 | C | 7 |
| qSNAb_LA7.1 | Env. 2 | S7_12220650 | A:T | 0.09 | 2E-05 | 0.26 | −0.56 | 3 | 2E-05 | 0.27 | −0.55 | 3 | A | 2 |
R2 indicates phenotypic variation of a trait accounted for by the cSNP (R2 of the SNP with model). The allelic effect (AE) is in respect to the minor allele and is estimated from the MLM implemented using the Genome Association and Prediction Integrated Tool. The marker rank is given based on P-values for the corresponding chromosome for each trait. FA (Favorable allele), indicates allele that is related to higher SD.
Indicates number of annotated genes around cSNP (±50 kb).
Indicates locus also detected using phenotypic data averaged across environments. The remaining unmarked loci are unique to specific environments. The details of genetic loci detected for phenotypic data averaged across environments (combined) are presented in Supplemental Table S3.
MAF, minor allele frequency.
GWAS identifies genomic regions associated with stomatal traits
Considering the significantly (P < 0.001) strong relationship between predicted and manual observations of Ab and Ad SD, only those loci that were identified using the manual phenotyping are discussed hereafter (Supplemental Table S3). In this report, we focus primarily on the Ab surface, and results for the Ad surface are given in Supplemental Table S3.
Genetic loci for SD
GWAS analysis revealed a total of 39 cSNPs associated with the four stomatal traits, including Ab SD (10 for SDAb, mm−2), Ad SD (7 for SDAd, mm−2), Ab stomatal number per leaf (8 for SNAb_LA), and Ad stomatal number per leaf (14 for SNAd_LA) across environments (Figure 4; Supplemental Table S3). Of the total loci identified for SD and number traits, 14 loci were common between combined and individual environment GWAS (Supplemental Table S3 and Supplemental Figures S7 and S8).
GWAS identified 10 loci for SDAb across environments, among which one locus (qSDAb2.2) was detected in both environments (Table 2; Figure 4). Among the 10 loci for Ab SD, 4 loci (qSDAb6.3, qSDAb2.2, qSDAb7.1, and qSDAb6.2) were also detected in the combined GWAS (Table 2; Supplemental Table S3). The cSNPs of loci qSDAb6.3 (S6_50424601, P = 4E-06) and qSDAb8.1 (S8_58766382, P = 7E-06) associated with Ab SD (mm−2) had the lowest P-values in Env. 1 and Env. 2, respectively (Table 2). Three loci (qSDAb6.1, qSDAb6.2, and qSDAb6.3) on chromosome 6 had a negative effect (small to medium) on SDAb. Conversely, the minor alleles of loci, qSDAb2.1 (A) and qSDAb10.1 (C) positively affected SDAb in Env. 1 (Table 2). Among the genetic loci identified for SDAb in Env. 1, FAs of cSNPs, including S2_61654537 (A), S6_50424601 (T), and S10_5625444 (C), had significantly higher Ab SD in both environments (Supplemental Figure S11A). Similarly, FAs of cSNPs, S6_48877403 (G), S7_58134055 (T), and S8_58766382 (C) identified in Env. 2 showed significantly higher Ab SD in both environments (Supplemental Figure S11B). All annotated genes within ±50 kb distance from the cSNPs were extracted to identify causative genes responsible for stomatal traits. Four (qSDAb7.1) to 18 (qSDAb6.2) genes were located in close proximity to the cSNPs (Table 2), and the relevance of these genes is highlighted in the discussion section. In brief, most of the identified genetic loci had genes related to cell division, growth-promoting factors, transcription factors, transporters, kinases, and antioxidants genes (Supplemental Table S5).
For Ad SD (mm−2), 7 genetic loci were detected either in Env.1 (qSDAd5.1, qSDAd3.1, qSDAd2.1, qSDAd8.1, qSDAd10.2, and qSDAd4.1) or Env. 2 (qSDAd10.1), having 37 and 8 annotated genes, respectively (Supplemental Table S3; Figure 4). The cSNP (S10_6848960) at qSDAd10.1 was common between Env. 2 and combined GWAS (Supplemental Table S3). Minor alleles of two loci (qSDAd8.1 and qSDAd10.2) had a negative effect on SDAd, whereas the minor allele of the remaining five loci had a positive effect on SDAd (Supplemental Table S3). For stomatal number per leaf (SN_LA, product of SD and leaf area), eight genetic loci (five in Env. 1 and three in Env. 2) were identified for Ab surface (Table 2). Two loci, qSNAb_LA8.1 in Env. 1 and qSNAb_LA3.2 in Env. 2, were also detected in the combined GWAS (Table 2; Supplemental Table S3). Four of eight loci (qSNAb_LA2.1, qSNAb_LA3.1, qSNAb_LA7.1, and qSNAb_LA10.1) had a negative effect on stomatal number on the Ab surface (Table 2). Furthermore, four loci (qSNAb_LA2.1, qSNAb_LA3.2, qSNAb_LA7.1, and qSNAb_LA8.1) associated with the Ab stomatal number per leaf were colocalized with the Ad stomatal number per leaf (qSNAd_LA7.1, qSNAd_LA3.2, qSNAd_LA7.1, and qSNAd_LA8.1, respectively; Figure 4; Supplemental Table S3).
Genetic loci for SCA
For SCA, the GWAS analysis revealed 8 genetic loci for Ab SCA (5 in Env. 1 and 3 in Env. 2; Table 3) and 10 for Ad SCA (4 in Env. 1 and 6 in Env. 2; Supplemental Table S3). Two loci, qSCAAb2.1 and qSCAAb5.1, were consistently detected for Ab SCA in Env. 1 and combined GWAS; one locus (qSCAAb2.2) was common between Env. 2 and combined GWAS (Table 3), suggesting that some loci are environment-specific (Table 3). The minor alleles of three cSNPs (S3_8085701, S5_67460377, and S10_57774713) had a negative effect on SCAAb, while the minor alleles of the other two loci had a positive effect in Env. 1 (Table 3). The cSNP S2_63821651 was found across environments for both Ab SCA (qSCAAb2.2 in Env. 2) and Ad SCA (qSCAAd2.1 in Env. 1), with the associated minor allele having a positive effect in both cases (Supplemental Table S3; Figure 4). There was one common cSNP (S10_47688264) between environments for Ad SCA (Supplemental Table S3).
Table 3.
Summary of cSNPs for Ab SCA (μm2) in Environments 1 and 2
| Locus | Environment | cSNP | Alleles | MAF | P-value | R2 | AE | FA | Genesa |
|---|---|---|---|---|---|---|---|---|---|
| qSCAAb2.1 | Env. 1b | S2_44645217 | T:G | 0.09 | 3E-06 | 0.24 | 38.2 | G | – |
| qSCAAb10.1 | Env. 1 | S10_48928696 | T:G | 0.08 | 2E-05 | 0.23 | 30.7 | G | 4 |
| qSCAAb3.1 | Env. 1 | S3_8085701 | T:C | 0.13 | 2E-05 | 0.23 | −26.3 | T | 9 |
| qSCAAb10.2 | Env. 1 | S10_57774713 | G:C | 0.38 | 3E-05 | 0.23 | −28.0 | G | 16 |
| qSCAAb5.1 | Env. 1b | S5_67460377 | T:G | 0.28 | 4E-05 | 0.22 | −25.5 | T | 17 |
| qSCAAb4.1 | Env. 2 | S4_1608257 | T:C | 0.30 | 1E-06 | 0.19 | 19.5 | C | 20 |
| qSCAAb1.1 | Env. 2 | S1_4463443 | T:G | 0.09 | 1E-06 | 0.19 | 30.6 | G | 17 |
| qSCAAb2.2 | Env. 2b | S2_63821651 | C:G | 0.42 | 4E-05 | 0.17 | 16.4 | G | 17 |
MAF indicates minor allele frequency. R2 indicates phenotypic variation of a trait accounted for by the cSNP (R2 of the SNP with model). The AE is in respect to the minor allele estimated from the MLM implemented using the Genome Association and Prediction Integrated Tool. FA (Favorable allele) that is related to larger SCA.
Indicates total number of annotated genes around cSNP (±50 kb).
Indicates locus also detected using phenotypic data averaged across environments. The remaining unmarked loci are unique to specific environments. The details of genetic loci detected for phenotypic data averaged across environments (combined) are presented in Supplemental Table S3.
Loci for SinLA
GWAS scan for SinLA resulted in identifying 14 genetic loci (9 in Env. 1 and 5 for Env. 2; Supplemental Figure S10). Among the 14 loci identified, two (qSinLA3.4 and qSinLA9.1) were consistently associated between environments (Supplemental Figure S10C). We detected six cSNPs that emerged in common between Env. 1 and combined GWAS; and three common between Env. 2 and combined GWAS for SinLA (Supplemental Table S3; Supplemental Figure S10). The cSNP (S3_62740410) was commonly associated with a SinLA in both environments and SNAd_LA in Env.1 (qSNAd_LA3.1). The minor allele of this cSNP (S3_62740410, “A”) had a negative effect on both traits (Supplemental Table S3). Likewise, another locus on chromosome 6 (S6_52304035) was commonly associated with SinLA (qSinLA6.1) and Ab SD per leaf (qSNAb_LA6.1) in Env. 1, with the minor allele (T) at this locus having a positive effect on both traits (Supplemental Table S3). Two other cSNPs commonly associated with SinLA and stomatal traits were S8_54788016 (qSinLA8.1 and qSCAAd8.2 in Env. 2) and S2_72127928 (qSinLA2.3 in Env. 2 and qSNAd_LA2.3 in Env. 1; Supplemental Table S3).
Phenotypic response of haplotypes with contrasting SD
Stomata on the Ab surface are more responsive than stomata on the Ad surface, and they play a major role in gas exchange in response to changes in the environment (Willmer and Fricker, 1996; Harrison et al., 2020). An examination of the correlation between PCs of SNPs and phenotypic traits revealed no strong subpopulation influence on Ab SD under both environments (Supplemental Table S6). Since cSNP S6_50424601 (qSDAb6.3) is the only region strongly associated with Ab SD (mm−2) and consistently detected in both analyses (Figure 5; Table 4; Supplemental Figure S7), we investigated the causal haplotypes for this locus using pairwise LD correlations. Haplotype analysis of the targeted region (65 kb) revealed four haplotypes, with significant differences in the mean Ab SD (Figure 5, C and D). On average, 73% of the accessions carrying the GGTGG haplotype (at 50.39–50.46 Mb; on chromosome 6) were associated with significantly higher (more) Ab SD (mm−2) compared with AACCT across environments (Figure 5; Table 4). Ab SD was strongly correlated between experiments (Exp. 1 versus Exp. 2) in Env. 1 (r = 0.58, P < 0.01) and Env. 2 (r = 0.72, P < 0.01). Accessions carrying contrasting haplotypes for Ab SD but with a similar SCA were identified and reconfirmed in Exp. 2 (Table 4). The stomatal area fraction (product of Ab SD and SCA) had a weak association with gs in Env. 1 (R2 = 0.07) and Env. 2 (R2 = 0.03).
Figure 5.
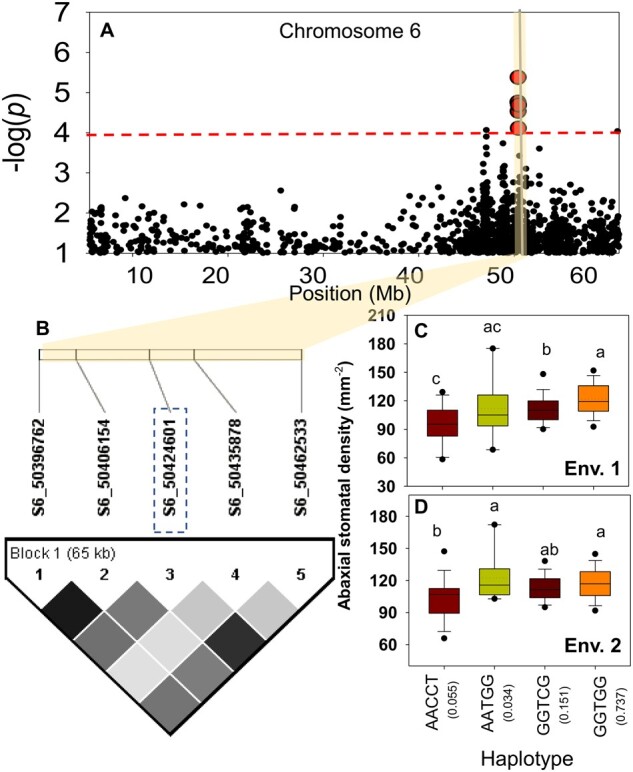
Regional plot of GWAS signal and pattern of pairwise LD (heatmap) for Ab SD per mm2 on chromosome 6. A, The −log10 (y-axis) of the P-values are plotted against their physical chromosomal position. The red dashed line indicates the significance threshold (−log10 = 4). The yellow bar indicates the most promising genomic region in Env. 1 selected for haplotype analysis. B, The LD heatmap was constructed using Haploview 4.2 software. The color intensity of the box corresponds with the r2 (light to dark gray indicate low to high recombination rate, respectively) between significant SNPs, S6_50396762, and S6_50462533. The SNP marked by dashed blue rectangle was the cSNP detected by MLM for Ab SD. C and D, SNPs highlighted in red in A are the five SNPs from which the haplotypes were formed. The whiskers indicate the interquartile range, and the outliers for Ab SD (mm−2) in Env. 1 (C) and Env. 2 (D). The dashed lines represent the mean, solid lines represent the median, and the whiskers indicate the 95% confidence interval. The haplotypes with a frequency (values in the parentheses) of >5% (AACCT, GGTCG, and GGTGG) were included for phenotypic reconfirmation (C and D). Means followed by a common letter are not significantly different by Tukey’s test at the 5% level of significance.
Table 4.
Accessions carrying the contrasting haplotype on chromosome 6 for Ab SD in Environments 1 and 2
| Haplotype | Accession | Ab SD (mm−2) |
Ab SCA (μm2) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 |
Experiment 2 |
Meana | Experiment 1 |
Experiment 2 |
Meana | ||||||
| Env. 1 | Env. 2 | Env. 1 | Env. 2 | Env. 1 | Env. 2 | Env. 1 | Env. 2 | ||||
| AACCT | PI598069 | 62.0 | 65.7 | 76.9 | 77.8 | 70.6 | 731.8 | 771.8 | 752.4 | 730.8 | 746.7 |
| AACCT | PI655983 | 85.2 | 76.4 | 94.4 | 90.7 | 86.7 | 765.4 | 788.1 | 719.9 | 832.4 | 776.4 |
| AACCT | PI653616 | 111.6 | 93.1 | 94.9 | 88.9 | 97.1 | 694.9 | 759.0 | 692.9 | 789.4 | 734.1 |
| AACCT | PI651496 | 96.3 | 110.2 | 126.4 | 106.9 | 110.0 | 774.2 | 657.2 | 682.1 | 770.0 | 720.9 |
| GGTCG | PI655995 | 94.0 | 94.9 | 92.6 | 83.8 | 91.3 | 705.8 | 711.0 | 720.7 | 773.3 | 727.7 |
| GGTCG | PI642998 | 109.7 | 108.3 | 113.0 | 107.9 | 109.7 | 644.1 | 702.2 | 616.3 | 634.6 | 649.3 |
| GGTCG | PI656036 | 116.2 | 108.3 | 159.3 | 84.7 | 117.1 | 762.7 | 809.3 | 684.2 | 903.8 | 790.0 |
| GGTGG | PI656107 | 115.3 | 95.8 | 124.1 | 91.2 | 106.6 | 847.6 | 833.1 | 789.1 | 943.5 | 853.3 |
| GGTGG | PI534075 | 117.1 | 97.2 | 114.8 | 112.0 | 110.3 | 761.4 | 725.0 | 734.9 | 676.9 | 724.6 |
| GGTGG | PI48770 | 108.8 | 113.0 | 118.5 | 108.3 | 112.2 | 706.5 | 689.2 | 648.8 | 704.3 | 687.2 |
| GGTGG | PI655972 | 114.8 | 112.0 | 121.8 | 111.1 | 114.9 | 714.4 | 646.6 | 677.2 | 708.7 | 686.7 |
| GGTGG | PI656003 | 134.3 | 116.2 | 118.5 | 118.1 | 121.8 | 724.1 | 648.4 | 639.8 | 680.1 | 673.1 |
| GGTGG | PI655976 | 135.7 | 113.9 | 136.1 | 106.0 | 122.9 | 672.0 | 724.5 | 692.5 | 802.6 | 722.9 |
| GGTGG | PI595745 | 128.2 | 146.3 | 123.6 | 100.9 | 124.8 | 667.3 | 609.3 | 733.9 | 740.8 | 687.8 |
| GGTGG | PI561472 | 139.4 | 111.1 | 141.7 | 110.2 | 125.6 | 705.9 | 743.9 | 706.5 | 799.4 | 738.9 |
| GGTGG | PI656065 | 142.6 | 105.1 | 124.1 | 132.4 | 126.0 | 727.4 | 719.3 | 707.8 | 632.5 | 696.8 |
| GGTGG | PI656015 | 134.7 | 119.0 | 142.6 | 120.4 | 129.2 | 720.5 | 722.1 | 718.5 | 724.3 | 721.3 |
| GGTGG | PI656074 | 142.6 | 128.2 | 124.1 | 124.1 | 129.7 | 624.9 | 700.8 | 669.2 | 693.3 | 672.0 |
| GGTGG | PI595739 | 142.6 | 128.2 | 153.2 | 110.2 | 133.6 | 775.1 | 814.1 | 776.8 | 862.2 | 807.1 |
| GGTGG | PI534101 | 146.3 | 137.5 | 152.8 | 112.5 | 137.3 | 729.1 | 761.4 | 729.2 | 835.8 | 763.9 |
| AACCT | 88.8a | 86.3a | 98.1a | 91.1a | 91.1a | 741.6a | 744.0a | 711.8a | 780.6a | 744.5a | |
| GGTCG | 106.6b | 103.9b | 121.6b | 92.1a | 106.1b | 704.2a | 740.8a | 673.7a | 770.6a | 722.3a | |
| GGTGG | 130.9c | 117.2c | 130.4b | 112.1b | 122.7c | 721.2a | 718.3a | 709.6a | 754.2a | 725.8a | |
Favorable haplotype associated with higher Ab SD is underlined.
Indicates averaged across two experiments. Accessions carrying contrasting haplotypes on chromosome 6 (at 50.39–50.46 Mb; qSDAb6.3) for Ab SD were identified using Haploview 4.2. Different lowercase letters indicate significant difference between haplotypes by one-way ANOVA.
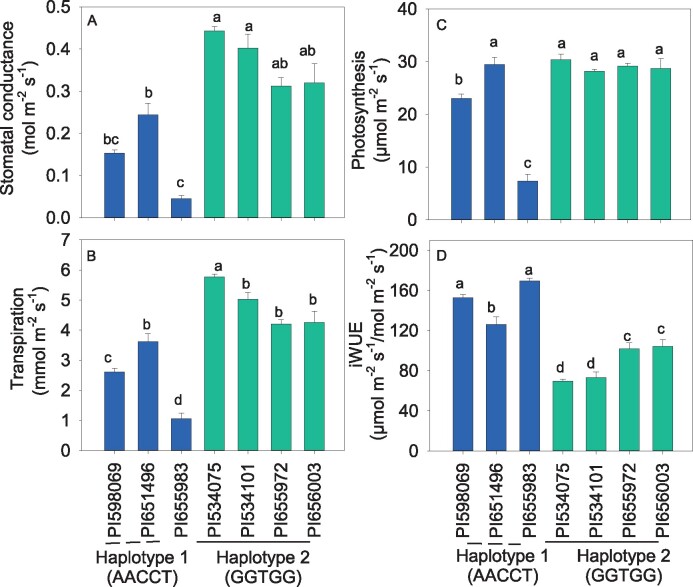
Further, gas exchange analysis was performed to characterize accessions with the most contrasting (favorable versus unfavorable; GGTGG versus AACCT) haplotypes of Ab SD and gs, but with a similar SCA under field conditions in Env. 1 (Figure 6, A–D; Supplemental Figure S12A). On average, selected accessions with an unfavorable haplotype (AACCT, fewer stomata) recorded significantly lower photosynthesis (28.3%), gs (57%), and transpiration (47%) compared with the favorable haplotype (GGTGG, denser stomata) accessions (Figure 6). Although the accession (PI651496) with AACCT haplotype recorded significantly lower gs (45%) and transpiration (37%) compared with PI534075 (Figure 6, A and B), it maintained a similar rate of photosynthesis as high SD accessions (Figure 6C). On the other hand, the intrinsic WUE (iWUE, the ratio of assimilation and gs) in the GGTGG haplotype was significantly lower (69%) than the accessions with AACCT (Figure 6D). As a result of large reduction in gs or transpiration and a relatively small reduction in assimilation, iWUE significantly increased in the PI651496 (Figure 6).
Figure 6.
Stomatal conductance (A), transpiration (B), photosynthesis (C), and iWUE (D) of sorghum accessions carrying contrasting haplotypes for Ab SD under field conditions. Bars ± se (n = 3). Means followed by a common letter are not significantly different by Tukey’s test at the 5% level of significance. Gas exchange parameters were measured on a fully opened young leaf for two days (66 and 67th days after planting) in Env. 1 under field conditions in Exp. 2.
Discussion
Classical phenotyping and deep learning methods unraveled the diversity in sorghum’s stomatal traits
Considerable evidence in field crops has shown the importance of stomatal characteristics and their association with photosynthesis and productivity (Farquhar and Sharkey, 1982), including rice (Ohsumi et al., 2007; Caine et al., 2019; Buckley et al., 2020), barley (Hughes et al., 2017), wheat (Dunn et al., 2019), and sorghum (Muchow and Sinclair, 1989). Previous studies have characterized the stomatal traits manually, either from a single environment or under controlled environments using limited genetic diversity, due to challenges associated with phenotyping. Phenotyping of diversity panels for stomatal traits following the classical approach is cumbersome, with reproducibility of results from large-scale studies posing a substantial bottleneck (Hudson, 2008; Furbank and Tester, 2011). To bridge this knowledge gap, we characterized the genetic diversity in sorghum stomatal traits from two different environments by developing and integrating deep learning-based high-throughput phenotyping (Figure 1). We targeted the middle portion of the second fully developed leaf from the top, which is known to have the highest SD at the 14 leaves stage in sorghum, to collect stomatal imprints (Liang et al., 1975). The integration of the automated deep learning method (https://github.com/matterport/Mask_RCNN) helped overcome the time-consuming manual method of stomata counting (Figure 2) and stomata complex area measurement (Figure 3), both in terms of speed and accuracy. Following the classical manual phenotyping approach, it took approximately 150 working days (∼3 min × 11, 196 images) to obtain the SD, while it took ∼7 d to obtain both SD and SCA by adopting the deep learning method.
In our study, both the Ab and Ad surfaces exhibited considerable and continuous variation in SD (Supplemental Figures S3 and S4) and SCA (Figure 3) among grain sorghum accessions (Table 1). On average, higher SD was recorded on the Ab surface (39% in Env. 1 and 32% in Env. 2) than the Ad, while the Ab SCA was lower (5% in Env. 1 and 2% in Env. 2) compared to the Ad surface. Similar variations in Ab and Ad SD have been reported in rice (Zhang et al., 2019; Chen et al., 2020). The Ab SD decreased by 2.5% (P < 0.01) in Env. 2 (low rainfall) compared to Env. 1 (high rainfall), but with a 7% (P < 0.001) increase in Ad SD in Env. 2 compared to Env. 1 (Supplemental Figure S3; Table 1). Reduced Ab or increased Ad SD in sorghum seems to be an adaptive response to low rainfall environments to minimize water loss, similar to other crops (Kondamudi et al., 2016). Decreased Ab SD (directly corresponds with reduced gs and water loss) is associated with improved WUE and drought tolerance in field crops (Hughes et al., 2017; Caine et al., 2019; Dunn et al., 2019). Increased Ad SD is proposed as a potential target to increase mesophyll conductance by creating a path for increased CO2 diffusion (Drake et al., 2019; Pathare et al., 2020) under drier conditions.
Candidate genetic loci associated with stomatal traits and their comparison with previous studies
This is the first report that has utilized the natural diversity in grain sorghum to characterize the genetic control of stomatal traits (SD and SCA) using GWAS, under field conditions (Figure 4). A comparison of GWAS results from manual and prediction-based datasets identified the same genetic loci (Table 2; Supplemental Table S2), indicating a robust, and highly efficient high-throughput deep learning method. Other companion studies in biomass sorghum (Ferguson et al., 2020) and maize (Xie et al., 2020) also show similar results. Our study revealed small to medium effect loci controlling the genetic architecture of stomatal traits in grain sorghum (Supplemental Table S3). This is in agreement with other studies in Arabidopsis (Dittberner et al., 2018) and in biomass sorghum (Ferguson et al., 2020) and maize (Xie et al., 2020).
Of the 71 genetic loci (38 were environment-specific and 33 common across environments) detected in the study, 46% of them were in close proximity or overlapped with previously reported genomic regions in different populations of sorghum (Figure 4; Supplemental Table S5). This allowed us to identify loci closely linked with other plant architectural traits in sorghum. For example, the qSDAb6.3 (at 50.4 Mb), controlling Ab SD in Env. 1, is within the (49.7–51.7 Mb) genomic region associated with number of green leaves and leaf width (Rama Reddy et al., 2014; Mccormick et al., 2016; Figure 4; Supplemental Table S4). Locus qSDAb2.1 overlaps with a previously identified locus for transpiration rate (Ortiz et al., 2017), reported in sorghum to be associated with gas exchange under chilling conditions (Figure 4; Supplemental Table S4). A locus, qSDAb7.1 overlapped with a quantitative trait locus (QTL) detected in sorghum for leaf width (Feltus et al., 2006) and grain yield in phosphorus-limited soils (Leiser et al., 2014; Figure 4; Supplemental Table S4). The qSDAb8.1 is proximal (0.35 Mb) to the genomic region associated with the efficiency of PSII center, and chlorophyll fluorescence of sorghum seedling under chilling stress (Fiedler et al., 2014; Ortiz et al., 2017). The qSDAb2.2 detected across environments (Table 2) colocalized with SD (Kapanigowda et al., 2014), leaf angle (Perez et al., 2014), stay-green, and chlorophyll content (Xu et al., 2000), suggesting that qSDAb2.2 could play an important role in leaf growth and development in sorghum (Figure 4; Supplemental Table S4). qSCAAb10.2 and qSNAb_LA6.1 were identified to overlap with the locus reported for transpiration ratio under drought stress in sorghum (Kapanigowda et al., 2014), indicating the pleiotropic effects of these loci. Interestingly, none of the identified cSNPs coincided with both Ab and Ad SD (mm−2), indicating that the genetic basis between the two leaf surfaces for stomatal traits may not be the same in sorghum (Supplemental Table S3). These findings warrant a systematic investigation of amphistomatal trait dynamics for optimizing carbon gain and water loss (Mott et al., 1982) to enhance WUE in sorghum under arid conditions. Converting the identified cSNPs, such as qSDAb6.3, qSDAb2.2, and qSCAAb10.2, into Kompetitive Allele-Specific PCR (polymerase chain reaction) markers and stacking them into a common genetic background would provide opportunities to manipulate SD in sorghum.
Promising candidate genes governing stomatal traits in the detected genomic regions
Genes governing traits related to stomatal traits and leaf development influence WUE in plants and serve as potential targets for trait-based breeding. Common genetic loci detected using classical and deep learning phenotyping methods resulted in discovering putative causal genes related to stomatal traits (Supplemental Table S5). As hypothesized, many genes related to leaf development, stomatal morphology, and development through phytohormone transport and signaling (auxin, abscisic acid [ABA], and brassinosteroids), stomatal lineage, phosphorylation, and subsequent degradation, and cell division, were found adjacent to loci associated with stomatal traits in sorghum (Supplemental Table S5), similar to other studies (Buckley et al., 2020; Chen et al., 2020; Zhu et al., 2020). Here, we present the most interesting loci and promising candidate genes for SD and SCA (Table 5).
Table 5.
Selected candidate genetic loci and potential candidate gene for SDAb and SCAAb in sorghum
| Locus | Environment | cSNP | FA | Gene ID | Sorghum bicolor annotation |
|---|---|---|---|---|---|
| Ab SD (mm−2) | |||||
| qSDAb2.2 a | 1, 2 | S2_61654537 | A | Sobic.002G224500 | bHLH factor, putative, expressed |
| qSDAb6.1 | 2 | S6_3637502 | A | Sobic.006G020700 | Homeobox and Steroidogenic acute regulatory protein (StAR)-related lipid transfer (START) domains containing protein |
| qSDAb6.2 a | 2 | S6_48877403 | G | Sobic.006G123100 | ABC transporter, ATP-binding protein |
| qSDAb6.3 a | 1 | S6_50424601 | T | Sobic.006G141700 | Glutaredoxin subgroup I (OsGrx_C2.2). Grx plays a critical role in protecting cells against oxidative stress damage |
| qSDAb7.1 a | 2 | S7_58134055 | T | Sobic.007G149700 | Similar to BRASSINOSTEROID-INSENSITIVE 1-associated receptor kinase 1, putative, expressed |
| qSDAb10.1 | 1 | S10_5625444 | C | Sobic.010G069600 | OsMKK5—putative mitogen-activated protein kinase based on amino acid sequence homology |
| qSDAb10.2 | 1 | S10_56551896 | C | Sobic.010G224400 | Potassium transporter-related protein |
| Ab SCA (μm2) | |||||
| qSCAAb3.1 a | 1 | S3_8085701 | T | Sobic.003G092700 | Xylosyltransferase, putative, expressed |
| qSCAAb4.1 a | 2 | S4_1608257 | C | Sobic.004G020600 | ARABIDOPSIS TRITHORAX-RELATED, putative, expressed |
| qSCAAb10.2 | 1 | S10_57774713 | G | Sobic.010G235400 | F-box, Leucine-rich repeats and Fibrin-binding domain containing proteins (OsFBLD1), expressed |
Indicates locus also detected using phenotypic data averaged across environments. The remaining unmarked loci are unique to specific environments. FA (Favorable allele) indicates allele that is related to denser stomata or large SCA.
Putative genes governing SD
A cSNP on chromosome 6 (S6_50424601; qSDAb6.3) for SDAb in Env. 1 was close (41.5 kb) to Sobic.006G141700, a homolog of rice Grx, glutaredoxin (Hu et al., 2017; Table 5). This gene is a homolog of glutaredoxin, an ubiquitous oxidoreductase that plays a substantial role in stress tolerance by reducing the rate of water loss via manipulating stomatal aperture in rice (Hu et al., 2017). Locus qSDAb6.2 associated with Ab SD in Env. 2, and combined GWAS found it was close (14 kb) to the ATP-binding cassette (ABC) transporter gene (Sobic.006G123100), which is a stomatal regulator (Kuromori et al., 2017). In the same study, Arabidopsis mutant, AtABCG22 (ABC G22) exhibited a drought susceptibility phenotype due to increased transpiration (Kuromori et al., 2017), which could be attributed to denser stomata. Likewise, in Env. 2, a locus qSDAb6.1 (3,637,502 bp), was associated with Ab SD and was found close (72 kb) to a putative homeodomain-START transcription factor gene (Sobic.006G020700). Transgenic plants involving the homeodomain-START gene in tobacco and Arabidopsis demonstrated enhanced drought tolerance via reduced SD and improved root architecture (Yu et al., 2008). Locus, qSDAb7.1 identified in Env. 2 and also in the combined analysis was close (46.5 kb from S7_58134055) to a gene (Sobic.007G149700) similar to BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1. BRASSINOSTEROID INSENSITIVE 1 (BRI1), a membrane-bound leucine-rich repeat receptor kinase (LRR-RK) is known to differentially modulate stomatal development. Studies have shown negative regulation by brassinosteroids on stomatal numbers (Kim et al., 2012). One locus (qSDAb10.1) on chromosome 10 (31.6 Mb) for Ab SD in Env. 1 is proximal (42 kb) to the mitogen-activated protein kinase family gene (Sobic.010G069600), which is known to negatively regulate stomatal development (Wang et al., 2007; Liu et al., 2010). Another locus (qSDAb10.2 in Env. 1) on chromosome 10 (at 56.6 Mb; Table 5) for Ab SD was close (70 kb) to a potassium transporter (Sobic.010G224400), which is known to be involved in regulating guard cell movement (an increase in K+ influx leads to stomatal opening), and thereby regulate transpiration in response to environmental signals (Schroeder et al., 2001). The locus qSDAb2.2, detected across environments, was positioned close (47.7 kb) to the basic helix–loop–helix family (bHLH, Sobic.002G224500) transcription factor, wherein its homolog is predicted to initiate stomatal development in Arabidopsis and grasses (Buckley et al., 2020). The bHLHs are also known as SPEECHLESS (SPCH), MUTE (switch for meristematic fate transition), FAMA (controls cell division and differentiation during stomatal development), INDUCER OF CBF1(ICE1)/SREAM1 (SCRM1 and SCRM2), modulating stomatal development processes (Buckley et al., 2020 and other references therein). Therefore, these genes have become notable targets for genetic manipulation of stomatal traits to improve plant productivity and stress tolerance.
Putative genes governing SCA
We detected a putative xylosyltransferase gene, Sobic.003G092700, near cSNP (25 kb; S3_8085701) for Ab SCA (qSCAAb3.1) in Env. 1 (Table 5). This gene is predicted to have a substantial role in coordinating the opening and closing of stomatal guard cells or regulating normal stomatal movement in Arabidopsis (Rui and Anderson, 2016). Interestingly, a locus (qSCAAb4.1) on chromosome 4 (1,608,257 bp) for the Ab SCA was beside (1.8 kb, Sobic.004G020600) the ARABIDOPSIS TRITHORAX-RELATED gene. A knockout mutant of ATX1 had larger stomatal apertures, increased water loss, and decreased tolerance to dehydration stress accompanied by decreased ABA accumulation, indicating the involvement of this gene in regulating the expression of genes associated with drought stress tolerance and ABA responses (Ding et al., 2011; Lee et al., 2017; Liu et al., 2018). cSNPs (S10_57774713; qSCAAb10.2 in Env. 1) at 57.7 Mb on chromosome 10, which was associated with variations in the Ab SCA, were seen to be physically located in the domain of LRR (Sobic.010G235400). A few well-known examples of LRR-RLKs are BRI1 (receptor for brassinosteroids) and ERECTA family LRR-RLKs, which are involved in controlling stomatal patterning, thus influencing transpiration efficiency in Arabidopsis (Meng et al., 2015). Future research focusing on these candidate loci and genes will advance our understanding and approach to improve the balance between water loss and carbon uptake in sorghum.
Optimizing stomatal traits to enhance WUE without yield penalty
Minimizing water loss by stomatal closure (stress avoidance) is an immediate response to limited water availability conditions, a strategy that has helped plants to adapt to a wide range of soil moisture conditions (Leakey et al., 2019). Under nonstress conditions, a leaf with large and few stomata is reported to result in higher WUE, accompanied by reduced photosynthesis rates, compared to plants with many smaller stomata on the leaf surface (Drake et al., 2013). Contrasting haplotypes for Ab SD phenotyped for gas exchange under field conditions helped identify accessions with low transpiration or gs and high photosynthesis (Figure 6, A and C). For example, under low rainfall arid regions, increased transpiration demand due to higher vapor pressure deficit reduces the leaf cell turgor and leaf size, affecting the carboxylation process, thereby limiting biomass accumulation and ultimately yield (Chaves et al., 2016). Therefore, under arid cropping regions, accessions with fewer stomata can be used as a selection strategy to manipulate gs and indirectly WUE (Muchow and Sinclair, 1989; Richards et al., 2002). Recently, wheat plants with 50% reduction in SD during tillering obtained via manipulation of Epidermal Patterning Factor (EPF) gene expression demonstrated increased iWUE without a significant reduction in yield under drought and elevated CO2, compared to control (Dunn et al., 2019). High-yielding cultivated rice varieties are inherently higher in gs than required. Hence, OsEPF1-overexpressed transgenic IR64 plants substantially reduced SD and gs under drought conditions, without a substantial reduction in grain yield (Caine et al., 2019). Similarly, reduced SD in Arabidopsis (Hara et al., 2009) and barley have been associated with enhanced WUE and drought tolerance (Hughes et al., 2017). Sorghum is grown under both irrigated and to a larger extent nonirrigated conditions. Therefore, the mechanism that allows a considerable reduction in gs (reducing transpiration) without affecting the maximum photosynthesis would be advantageous under water-limited environments. With these assumptions, we hypothesize that sorghum genotypes with fewer Ab stomata (resulting in lower gs and transpiration) with efficient CO2 assimilation would be an ideal choice to improve adaptation and WUE in stressful environments. When irrigation is not a limitation, choosing a genotype with denser stomata (thus, high gs and transpiration) would increase CO2 assimilation and biomass, including nutrient uptake. Therefore, manipulating genes controlling stomatal traits presents a practical alternative approach for improving WUE (Franks et al., 2015) and drought tolerance without affecting yield in sorghum. Our findings highlighted genetic loci and potential candidate genes associated with variation in SD and SCA, providing opportunities to explore mechanisms and optimize stomatal traits to improve adaptation in dryland crops.
In conclusion, we have developed an image-based high throughput deep learning tool to identify, classify, and record SD and area in sorghum. A 98% accuracy in predicting SD between manual and deep learning methods presents an excellent platform for other crops/plants to explore stomatal diversity for further enhancing crop adaptation under water-limited conditions. GWAS results from manual and prediction-based datasets indicated the reliability and efficiency of the deep learning method. Our findings have unlocked the genetic information housed in the sorghum genome that controls stomatal traits (using GWAS) and demonstrated the possibility of replacing laborious traditional stomata counting with an efficient and automated approach such as Mask R-CNN. Identified donor lines with favorable genetic loci are potential sources for stomata-targeted breeding and exploring molecular mechanisms that control stomatal regulation in sorghum to enhance adaptation under arid conditions with minimal to no yield penalty.
Materials and methods
Plant material and environments
The SAP consisting of 311 accessions was assembled from 25 countries representing major sorghum growing regions of the world (Casa et al., 2008; Morris et al., 2013). The SAP consisted of five grain sorghum races, (namely caudatum, bicolor, guinea, durra, and kafir), intermediate races, converted lines, and elite accessions of historical and geographic importance (Harlan and Wet, 1972). In experiment 1 (Exp. 1 in 2017), the SAP was grown in two different environments (Env. 1: Kansas State University, North Farm, Manhattan and Env. 2: Agricultural Research Centre at Hays, Kansas) in a randomized block design with two replications per accession per environment. All 311 accessions were planted in a single row plot of 6.1-m long, with 0.7-m spacing between rows. Approximately 50 seeds were sown per row for each accession. Three representative plants in the middle of the row, for each accession, were tagged for studying the natural variation in SD, SCA and SinLA. All measurements were recorded 62 d after planting in both environments (Env. 1 and 2). In experiment 2 (Exp. 2 in 2018), to reconfirm the expression of the trait, candidate accessions carrying the contrasting allelic haplotypes for Ab SD with similar SCA were planted in the same environments (Env. 1 and Env. 2) in 2018. Sixty-eight days after planting, we measured gs, the effective quantum yield (QY) of PS II, including the SD, SCA, and SinLA. The crop management and protocol for obtaining stomatal imprints and other data were the same across both experiments as detailed below. A schematic overview of the study is visualized in Figure 1 and the environmental conditions during the growing seasons at these locations are given in Supplemental Figure S1, A–C (2017-Exp. 1) and Supplemental Figure S1D–F (2018-Exp. 2).
Experiment 1
Stomatal density
To capture the natural variation in stomatal number and SCA, the Ab and Ad leaf surfaces were carefully smeared with a thin layer of transparent nail polish in the mid-portion of the fully opened leaf. Care was taken to identify the second leaf from the top that was fully open and completely developed, from which the imprints were obtained. After 3–5 min, thin imprints (∼25 × 17 mm2) were peeled off from both the leaf surfaces using tape (Scotch Transparent Clear Tape), and mounted on glass slides (75 × 25 mm2) following the procedure of Rowland-Bamford et al. (1990). Three random field of view images per slide were captured at ×400 magnification using the compound microscope (Olympus BX51 with DP 70 camera). From each image, the number of stomata was counted and divided by 0.24 mm2 (area of each field) to estimate SD. In brief, number of stomata (N) was manually counted per field of view (S = πr2, r = view radius) and SD was estimated as N/S (N = number of stomata mm−2), as described in another study (Drake et al., 2013). A total of 11,196 images (311 accessions × 3 plants × 2 environments × 2 leaf surfaces × 3 images per slide) were used to record stomatal traits (Table 1). Three leaves that were used for taking stomata imprints were harvested separately to determine the SinLA, using a leaf area meter (LI-3000; LI-COR, Lincoln, Nebraska, USA). Later, stomatal number per leaf was estimated to normalize the density on a whole leaf area basis, using the Ad and Ab SD per mm2 (Table 1).
An automated technique to predict SD and SCA
We used a fully automated deep learning method, called Mask R-CNN, to perform stomata instance segmentation for each input image, i.e. to identify the pixels corresponding to stomata in an image. Mask R-CNN (Figure 1C) is an extension of the Faster R-CNN approach (Ren et al., 2015). Similar to the Faster R-CNN network, Mask R-CNN can be trained to detect objects of interest (e.g. stomata) in an image, and to localize the objects detected using bounding boxes. In addition, Mask R-CNN generates a precise segmentation mask for each object instance. The Faster R-CNN network has two main components, which share a backbone feature extractor CNN, such as ResNet (He et al., 2016). The first component, called a Region Proposal Network (RPN), uses the last feature map produced by the backbone CNN to identify regions of interest (RoI), i.e. fragments of the image (called anchors) that may contain target objects and initial approximate bounding boxes for those objects. The second component consists of fully connected layers that classify RoI proposed by the RPN network into specific categories (an object classification task) and refine the corresponding bounding box coordinates (a bounding box regression task). Mask R-CNN extends the Faster R-CNN network by including additional convolutional layers trained to predict instance masks for RoI (an instance segmentation task), in parallel with the object classification and bounding box regression tasks. Furthermore, Mask R-CNN uses a Feature Pyramid Network (FPN; Lin et al., 2017) together with ResNet as the architectural backbone to enable the identification of objects at different scales. It also replaces the RoI Pool layer in Faster R-CNN, which extracts a fixed-length feature vector from a feature map, with a RoI Align layer, which performs pixel-to-pixel alignment between network input and output, to enable the generation of precise instance masks. We used the implementation of Mask R-CNN, available at https://github.com/matterport/Mask_RCNN, with ResNet101 as the backbone network (together with FPN). We changed the original Mask R-CNN architecture to customize it to our categories (stomata and background_ used for the object classification and instance segmentation tasks. The pretrained Mask R-CNN network was fine-tuned on datasets of increasingly larger sizes (specifically, 20, 50, 100, 200, and 300 images) and validated on a separate dataset consisting of 300 images. Using the training and validation loss curves, we selected the model trained on 300 images (280 images from Exp. 1 and 20 images from Exp. 2) to perform the stomata instance segmentation on the remaining images (i.e. images not included in the training and validation subsets), and subsequently produced the predictions (i.e. deep learning dataset) used in this study. All images used for training and validation had stomata labeled using the VGG Image Annotator (1.0.6) tool, available at http://www.robots.ox.ac.uk/∼vgg/software/via/. The number of stomata in an image was obtained from the segmentation result and used to calculate SD, which was compared to the density obtained based on manual counting. Subsequently, the instance masks were used to calculate SCA, and the results were validated based on 50 images where stomatal area was manually measured using ImageJ (https://imagej.nih.gov/ij/). Finally, SCA was calculated for all images from Exp. 1 and Exp. 2 using the predicted stomata masks.
Phenotypic data analyses
All the phenotypic traits collected were analyzed using analysis of variance (ANOVA) to test the effect of genotype (G), environment (E), and their interaction using GenStat (18th Edition, http://www.vsni.co.uk). The PCA was performed in XLSTAT. The chart.Correlation () function within the R package “Performance Analytics” was used to generate the correlation scatter plot. The H2 of all the measured traits was estimated considering the proportion of phenotypic variance that is due to the genetic variance.
GWAS analyses
A total of 308 accessions had genotypic and complete phenotypic data in both environments, hence GWAS was performed using 308 accessions. The genotype by sequencing SNP information for the panel has been described by Morris et al. (2013). From the above, 184,002 SNPs were used for GWAS analyses after filtering for minor allele frequency (5%) using TASSEL 5.2.3 (Bradbury et al., 2007). A MLM was performed on both manual and deep learning datasets to identify loci associated with target traits (Lipka et al., 2012) using Genome Association and Prediction Integrated Tool (http://www.zzlab.net/GAPIT/). GWAS was performed using the first three principal components (PCA.total = 3) as covariates (which adequately explained the population structure) and with the default individual genetic relatedness matrix (K) based on VanRaden method, as described previously for the same population (Moghimi et al., 2019). We did not find any significant SNPs at a threshold above the Bonferroni corrected P-value of 0.05 (−log10(0.05/1,84,002)=6.57). Therefore, we chose a suggestive threshold of −log10 ≥ 4 to detect the significant marker trait associations, as followed recently for the same population (Moghimi et al., 2019). Manhattan and Q−Q plots were generated using the library(qqman) in RStudio 3.6.1. Furthermore, to prioritize genomic regions associated with traits, importance was given to the SNP with the lowest P-value detected in both datasets. With the reported LD decay background level of up to 150 kb (Morris et al., 2013; Ortiz et al., 2017; Moghimi et al., 2019), the SNP with the lowest P-value within the 100 kb was considered as a cSNP to represent that locus. GWAS results from the current and previous studies were compared by extracting the genomic regions associated with similar or closely related traits including leaf morphology, stay green, and yield traits (Supplemental Table S4). A list of previously reported genomic regions (loci underlined in black in Figure 4) was obtained from https://aussorgm.org.au/sorghum-qtl-atlas/ (Mace et al., 2019). Furthermore, annotated genes around 100 kb of the cSNPs (±50 upstream and downstream of the cSNP) were extracted by scanning version 3.1 of the Sorghum bicolor genome.
Experiment 2
To identify donor accessions for physiological studies, haplotypes for the significant peak at qSDAb6.3 (S6_50424601) were identified using Haploview 4.2. Phenotypic means of each haplotype were calculated as the average of all accessions belonging to a specific haplotype (Figure 5). Haplotypes that appeared in >5% of the accessions were considered to test differences in a phenotype. Differences in mean phenotypic values of Ab SD haplotypes were tested by one-way ANOVA and Tukey’s test to select the contrasting haplotypes. Furthermore, 20 accessions carrying contrasting haplotypes (for Ab SD with a similar SCA) on chromosome 6 (at 50.39–50.46 Mb; qSDAb6.3) were identified (Table 4) and phenotyped in 2018 in both Env. 1 and Env. 2 (Exp. 2) similar to 2017. The crop management and protocol of data collection and image analysis methods were same as in Exp. 1. In this experiment, we measured gs, the effective QY of PS II in both environments, including all other parameters similar to Exp. 1. gs measurements were taken from three plants per accession on the mid-portion of the fully developed leaf for 3 d between 11:00 and 14:00 h, using a calibrated leaf porometer (Decagon Devices, Inc., Pullman, WA, USA). Similar to gs, light adapted QY measurements were taken on the same leaf using a portable fluorometer FluorPen (FluorPen FP 100, Photon System Instruments Ltd., Brno, Czech Republic).
Gas exchange parameters
Accessions with contrasting Ab stomatal densities were selected to test the functional relevance of SD in regulating gas exchange parameters in Env. 1. The gas exchange measurements were recorded on the mid-portion of the fully opened first leaf from the top (n = 3 plants per accession for 2 d) using a portable infrared gas analyzer (LI-6400XT, LI-COR, Lincoln, Nebraska, USA). Measurements were taken under constant leaf temperature of 30°C, PPFD (Photosynthetic Photon Flux Density) at 1,500 μmol m−2 s−1, relative humidity at ∼70%, and CO2 concentration at 400 μmol m−2 s−1 (carbon dioxide was supplied using external CO2 cartridges) between 10:00 and 13:00 h on bright sunny days. The iWUE was calculated as the ratio of the net photosynthetic rate (A) and the stomatal conductance (gs).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Environmental conditions observed during the experimental period.
Supplemental Figure S2. Comparison of models trained with different dataset sizes.
Supplemental Figure S3. Phenotypic distribution of abaxial and adaxial stomatal density captured using classical phenotyping method.
Supplemental Figure S4. Phenotypic distribution of abaxial and adaxial stomatal density captured using deep learning method.
Supplemental Figure S5. PCA plots for all manual and predicted datasets across environments.
Supplemental Figure S6. Pearson’s correlation matrix of phenotypic traits.
Supplemental Figure S7 Manhattan plots displaying GWAS results for abaxial stomatal density (per mm2) and number (per leaf) using a mixed linear model in two environments.
Supplemental Figure S8. Manhattan plots displaying GWAS results for adaxial stomatal density (per mm2) and number (per leaf) using a mixed linear model in two environments.
Supplemental Figure S9. Manhattan plots displaying genome-wide association results for predicted abaxial and adaxial stomatal complex area.
Supplemental Figure S10. Manhattan plots displaying GWAS results for single leaf area using a mixed linear model in two environments.
Supplemental Figure S11. Contribution of major and minor alleles to abaxial stomatal density in two environments.
Supplemental Figure S12. Stomatal conductance and QY of PSII of selected accessions across three days in environment 1 and 2.
Supplemental Table S1. A list of SAP accessions used in the study.
Supplemental Table S2. A list of identified genome-wide significant association loci for stomatal density between manual and deep learning methods.
Supplemental Table S3. Summary of identified cSNPs for phenotypic traits across environments.
Supplemental Table S4. List of previously reported genomic regions associated with similar or closely related traits in sorghum.
Supplemental Table S5. Survey of genes around each of the cSNP detected in the study.
Supplemental Table S6. Correlation (r) between the first three principal components (PCs) of SNPs and phenotypic traits.
Supplementary Material
Acknowledgments
Contribution number 20-316-J from the Kansas Agricultural Experiment Station. We thank Crop Ecophysiology (Manhattan) and Agricultural Research Center (Hays) research teams, Kansas State University for assistance with sample and data collection.
Funding
This work was not funded.
Conflict of interest statement. Authors declare no conflict of interest.
R.B., R.P., D.C. and S.V.K.J. designed and implemented the experiments; R.B., A.C., M.P., N.M., E.B., and T.O. collected the observational data; C.W. and D.C. performed the deep learning analysis; R.B. analyzed observational data and performed Genome-Wide Association Study; R.B., D.C., and S.V.K.J. interpreted the data and wrote the article. All authors contributed towards finalizing the manuscript. S.V.K.J. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: S.V. Krishna Jagadish (kjagadish@ksu.edu).
References
- Anderson VJ, Briske DD (1990) Stomatal distribution, density and conductance of three perennial grasses native to the southern true prairie of Texas. Am Midl Nat 123: 152–159 [Google Scholar]
- Bertolino LT, Caine RS, Gray JE (2019) Impact of stomatal density and morphology on water-use efficiency in a changing world. Front Plant Sci 10: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Buckley CR, Caine RS, Gray JE (2020) Pores for thought: can genetic manipulation of stomatal density protect future rice yields? Front Plant Sci 10: 1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, et al. (2019) Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JE, Adams CA, Holsinger KE (2016) Intraspecific variation in stomatal traits, leaf traits and physiology reflects adaptation along aridity gradients in a South African shrub. Ann Bot 117: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48: 30–40 [Google Scholar]
- Casado Á, Heras J (2018) Guiding the creation of deep learning-based object detectors. arXiv:1809.03322 (Accessed on April 7, 2021)
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ (2011) Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr Biol 21: 1025–1029 [DOI] [PubMed] [Google Scholar]
- Chater CCC, Caine RS, Fleming AJ, Gray JE (2017) Origins and evolution of stomatal development. Plant Physiol 174: 624–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Costa JM, Zarrouk O, Pinheiro C, Lopes CM, Pereira JS (2016) Controlling stomatal aperture in semi-arid regions—The dilemma of saving water or being cool? Plant Sci 251: 54–64 [DOI] [PubMed] [Google Scholar]
- Chen H, Zhao X, Zhai L, Shao K, Jiang K, Shen C, Chen K, Wang S, Wang Y, Xu J (2020) Genetic bases of the stomata-related traits revealed by a genome-wide association analysis in rice (Oryza sativa L.). Front Genet 11: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M (2011) The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways: ATX1 functions in dehydration stress responses. Plant J 66: 735–744 [DOI] [PubMed] [Google Scholar]
- Dittberner H, Korte A, Mettler-Altmann T, Weber APM, Monroe G, de Meaux J(2018) Natural variation in stomata size contributes to the local adaptation of water-use efficiency in Arabidopsis thaliana. Mol Ecol 27: 4052–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny-Adams T, Hunt L, Franks PJ, Beerling DJ, Gray JE (2012) Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philos Trans R Soc Lond B Biol Sci 367: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC, Berry JA (2014a) An integrated model of stomatal development and leaf physiology. New Phytol 201: 1218–1226 [DOI] [PubMed] [Google Scholar]
- Dow GJ, Berry JA, Bergmann DC (2014b) The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsis thaliana. New Phytol 201: 1205–1217 [DOI] [PubMed] [Google Scholar]
- Drake PL, Boer HJ, Schymanski SJ, Veneklaas EJ (2019) Two sides to every leaf: water and CO 2 transport in hypostomatous and amphistomatous leaves. New Phytol 222: 1179–1187 [DOI] [PubMed] [Google Scholar]
- Drake PL, Froend RH, Franks PJ (2013) Smaller, faster stomata: scaling of stomatal size, rate of response, and stomatal conductance. J Exp Bot 64: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte KTN, Carvalho MAG, de, Martins PS(2017) Segmenting high-quality digital images of stomata using the wavelet spot detection and the watershed transform. Proceedings of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications. SCITEPRESS - Science and Technology Publications, Porto, Portugal, pp 540–547 [Google Scholar]
- Dunn J, Hunt L, Afsharinafar M, Meselmani MA, Mitchell A, Howells R, Wallington E, Fleming AJ, Gray JE (2019) Reduced stomatal density in bread wheat leads to increased water-use efficiency. J Exp Bot 70: 4737–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanourakis D, Giday H, Milla R, Pieruschka R, Kjaer KH, Bolger M, Vasilevski A, Nunes-Nesi A, Fiorani F, Ottosen C-O (2015) Pore size regulates operating stomatal conductance, while stomatal densities drive the partitioning of conductance between leaf sides. Ann Bot 115: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli M, Matthews J, Lawson T (2019) Exploiting natural variation and genetic manipulation of stomatal conductance for crop improvement. Curr Opin Plant Biol 49: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD, Sharkey TD (1982) Stomatal conductance and photosynthesis. Annu Rev Plant Physiol 33: 317–345 [Google Scholar]
- Feltus FA, Hart GE, Schertz KF, Casa AM, Kresovich S, Abraham S, Klein PE, Brown PJ, Paterson AH (2006) Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor Appl Genet 112: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Ferguson JN, Fernandes SB, Monier B, Miller ND, Allan D, Dmitrieva A, Schmuker P, Lozano R, Valluru R, Buckler ES et al. (2020). Machine learning enabled phenotyping for GWAS and TWAS of WUE traits in 869 field-grown sorghum accessions. Plant Biol bioRxiv. 10.1101/2020.11.02.365213 (Accessed on April 7, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter KC, Eberhardt S, Barclay RS, Wing S, Keller SR (2019) StomataCounter: a neural network for automatic stomata identification and counting. New Phytol 223: 1671–1681 [DOI] [PubMed] [Google Scholar]
- Fiedler K, Bekele WA, Duensing R, Gründig S, Snowdon R, Stützel H, Zacharias A, Uptmoor R (2014) Genetic dissection of temperature-dependent sorghum growth during juvenile development. Theor Appl Genet 127: 1935–1948 [DOI] [PubMed] [Google Scholar]
- Field C, Merino J, Mooney HA (1983) Compromises between water-use efficiency and nitrogen-use efficiency in five species of California evergreens. Oecologia 60: 384–389 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Beerling DJ (2009) Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc Natl Acad Sci USA 106: 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD (2007) The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol 143: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Doheny-Adams TW, Britton-Harper ZJ, Gray JE (2015) Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol 207: 188–195 [DOI] [PubMed] [Google Scholar]
- Furbank RT, Tester M (2011) Phenomics – technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Gitz DC, Baker JT (2009) Methods for creating stomatal impressions directly onto archivable slides. Agron J 101: 232–236 [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Harlan JR, Wet JMJ (1972) A simplified classification of cultivated Sorghum 1. Crop Sci 12: 172–176 [Google Scholar]
- Harrison EL, Arce Cubas L, Gray JE, Hepworth C (2020) The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J 101: 768–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, Las Vegas, NV, USA, pp 770–778
- Henry C, John GP, Pan R, Bartlett MK, Fletcher LR, Scoffoni C, Sack L (2019) A stomatal safety-efficiency trade-off constrains responses to leaf dehydration. Nat Commun 10: 3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C, Caine RS, Harrison EL, Sloan J, Gray JE (2018) Stomatal development: focusing on the grasses. Curr Opin Plant Biol 41: 1–7 [DOI] [PubMed] [Google Scholar]
- Hepworth C, Doheny-Adams T, Hunt L, Cameron DD, Gray JE (2015) Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol 208: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Hasezawa S (2014) CARTA-based semi-automatic detection of stomatal regions on an Arabidopsis cotyledon surface. Plant Morphol 26: 9–12 [Google Scholar]
- Hu Y, Wu Q, Peng Z, Sprague SA, Wang W, Park J, Akhunov E, Jagadish KSV, Nakata PA, Cheng N, et al. (2017) Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci Rep 7: 15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME (2008) Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol Ecol Resour 8: 3–17 [DOI] [PubMed] [Google Scholar]
- Hughes J, Hepworth C, Dutton C, Dunn JA, Hunt L, Stephens J, Waugh R, Cameron DD, Gray JE (2017) Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol 174: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakody H, Liu S, Whitty M, Petrie P (2017) Microscope image based fully automated stomata detection and pore measurement method for grapevines. Plant Methods 13: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanigowda MH, Payne WA, Rooney WL, Mullet JE, Balota M (2014) Quantitative trait locus mapping of the transpiration ratio related to preflowering drought tolerance in sorghum (Sorghum bicolor). Funct Plant Biol 41: 1049. [DOI] [PubMed] [Google Scholar]
- Kawamitsu Y, Agata W, Hiyane S, Murayama S, Nose A, Shinjyo C (1996) Relation between leaf gas exchange rate and stomate. i. stomatal frequency and guard cell length in C3 and C4 grass species. Jpn J Crop Sci 65: 626–633 [Google Scholar]
- Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, co2, and ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Michniewicz M, Bergmann DC, Wang Z-Y (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue S, et al. (2011) FLOWERING LOCUS T regulates stomatal opening. Curr Biol 21: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Kondamudi R, Swamy K, Rao YV, Kiran TV, Suman K, Rao DS, Rao PR, Subrahmanyam D, Sarla N, Kumari BR (2016) Gas exchange, carbon balance and stomatal traits in wild and cultivated rice (Oryza sativa L.) genotypes. Acta Physiol Plant 38: 160 [Google Scholar]
- Kuromori T, Sugimoto E, Ohiraki H, Yamaguchi-Shinozaki K, Shinozaki K (2017) Functional relationship of AtABCG21 and AtABCG22 in stomatal regulation. Sci Rep 7: 12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laga H, Shahinnia F, Fleury D (2014) Image-based plant stomata phenotyping. 13th International Conference on Control Automation Robotics & Vision (ICARCV). IEEE, Singapore, pp 217–222 [Google Scholar]
- Lawson T, Blatt MR (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164: 1556–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, James W, Weyers J (1998) A surrogate measure of stomatal aperture. J Exp Bot 49: 1397–1403 [Google Scholar]
- Leakey ADB, Ferguson JN, Pignon CP, Wu A, Jin Z, Hammer GL, Lobell DB (2019) Water use efficiency as a constraint and target for improving the resilience and productivity of c 3 and c 4 crops. Annu Rev Plant Biol 70: 781–808 [DOI] [PubMed] [Google Scholar]
- Lee K, Park O-S, Seo PJ (2017) Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci Signal 10: eaan0316. [DOI] [PubMed] [Google Scholar]
- Leff B, Ramankutty N, Foley JA (2004) Geographic distribution of major crops across the world: Global crop distribution. Global Biogeochem Cycles 18: GB1009 [Google Scholar]
- Leiser WL, Rattunde HFW, Weltzien E, Cisse N, Abdou M, Diallo A, Tourè AO, Magalhaes JV, Haussmann BI (2014) Two in one sweep: aluminum tolerance and grain yield in P-limited soils are associated to the same genomic region in West African Sorghum. BMC Plant Biol 14: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang GH, Dayton AD, Chu CC, Casady AJ (1975) Heritability of stomatal density and distribution on leaves of grain sorghum 1. Crop Sci 15: 567–570 [Google Scholar]
- Lin T-Y, Dollar P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, Honolulu, HI, pp 936–944. [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang A, Yin H, Meng Q, Yu X, Huang S, Wang J, Ahmad R, Liu B, Xu Z-Y (2018) Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol 217: 1582–1597 [DOI] [PubMed] [Google Scholar]
- Liu Y-K, Liu Y-B, Zhang M-Y, Li D-Q (2010) Stomatal development and movement: the roles of MAPK signaling. Plant Signal Behav 5: 1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace E, Innes D, Hunt C, Wang X, Tao Y, Baxter J, Hassall M, Hathorn A, Jordan D (2019) The Sorghum QTL Atlas: a powerful tool for trait dissection, comparative genomics and crop improvement. Theor Appl Genet 132: 751–766 [DOI] [PubMed] [Google Scholar]
- Mccormick RF, Truong SK, Mullet JE (2016) 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros DB, Martins SCV, Cavalcanti JHF, Daloso DM, Martinoia E, Nunes-Nesi A, DaMatta FM, Fernie AR, Araújo WL (2016) Enhanced photosynthesis and growth in atquac1 knockout mutants are due to altered organic acid accumulation and an increase in both stomatal and mesophyll conductance. Plant Physiol 170: 86–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghimi N, Desai JS, Bheemanahalli R, Impa SM, Vennapusa AR, Sebela D, Perumal R, Doherty CJ, Jagadish SVK (2019) New candidate loci and marker genes on chromosome 7 for improved chilling tolerance in sorghum. J Exp Bot 70: 3357–3371 [DOI] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott KA, Gibson AC, O’Leary JW (1982) The adaptive significance of amphistomatic leaves. Plant Cell Environ 5: 455–460 [Google Scholar]
- Muchow RC, Sinclair TR (1989) Epidermal conductance, stomatal density and stomatal size among genotypes of Sorghum bicolor (L.) Moench. Plant Cell Environ 12: 425–431 [Google Scholar]
- Ohsumi A, Kanemura T, Homma K, Horie T, Shiraiwa T (2007) Genotypic variation of stomatal conductance in relation to stomatal density and length in rice ( Oryza sativa L.). Plant Prod Sci 10: 322–328 [Google Scholar]
- Ortiz D, Hu J, Salas Fernandez MG (2017) Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J Exp Bot 68: 4545–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CP, Freckleton RP (2009) Ecological selection pressures for C4 photosynthesis in the grasses. Proc R Soc B 276: 1753–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare VS, Koteyeva N, Cousins AB (2020) Increased adaxial stomatal density is associated with greater mesophyll surface area exposed to intercellular air spaces and mesophyll conductance in diverse C4 grasses. New Phytol 225: 169–182 [DOI] [PubMed] [Google Scholar]
- Pearce DW, Millard S, Bray DF, Rood SB (2006) Stomatal characteristics of riparian poplar species in a semi-arid environment. Tree Physiol 26: 211–218 [DOI] [PubMed] [Google Scholar]
- Perez MMB, Zhao J, Yin Y, Hu J, Salas Fernandez MG (2014) Association mapping of brassinosteroid candidate genes and plant architecture in a diverse panel of Sorghum bicolor. Theor Appl Genet 127: 2645–2662 [DOI] [PubMed] [Google Scholar]
- Rama Reddy N, Ragimasalawada M, Sabbavarapu M, Nadoor S, Patil J (2014) Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35-1 and a popular stay-green genotype B35. BMC Genomics 15: 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA (2014) Speedy small stomata? J Exp Bot 65: 1415–1424 [DOI] [PubMed] [Google Scholar]
- Reich PB (1984) Leaf stomatal density and diffusive conductance in three amphistomatous hybrid poplar cultivars. New Phytol 98: 231–239 [Google Scholar]
- Ren S, He K, Girshick R, Sun J (2015) Faster R-CNN: towards real-time object detection with region proposal networks. Proceedings of the 28th International Conference on Neural Information Processing Systems, Vol. 1. MIT Press, Cambridge, MA, USA, pp 91–99. [Google Scholar]
- Richards RA, Rebetzke GJ, Condon AG, van Herwaarden AF (2002) Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci 42: 111. [DOI] [PubMed] [Google Scholar]
- Rowland-Bamford AJ, Nordenbrock C, Baker JT, Bowes G, Hartwell Allen L (1990) Changes in stomatal density in rice grown under various CO2 regimes with natural solar irradiance. Environ Exp Bot 30: 175–180 [Google Scholar]
- Rui Y, Anderson CT (2016) Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol 170: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoda K, Watanabe T, Sukemura S, Kobayashi S, Nagasaki Y, Tanaka Y, Shiraiwa T (2019) Genetic diversity in stomatal density among soybeans elucidated using high-throughput technique based on an algorithm for object detection. Sci Rep 9: 7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T (2007) Light regulation of stomatal movement. Annu Rev Plant Biol 58: 219–247 [DOI] [PubMed] [Google Scholar]
- Tack J, Lingenfelser J, Jagadish SVK (2017) Disaggregating sorghum yield reductions under warming scenarios exposes narrow genetic diversity in US breeding programs. Proc Natl Acad Sci USA 114: 9296–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Fujii K, Shiraiwa T (2010) Variability of leaf morphology and stomatal conductance in soybean [Glycine max (L.) Merr.] cultivars. Crop Sci 50: 2525–2532 [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer C, Fricker M (1996) Stomata. Springer, Netherlands, Dordrecht, pp 126–191 [Google Scholar]
- Xie J, Mayfield-Jones D, Erice G, Choi M, Leakey ADB. 2020. Optical topometry and machine learning to rapidly phenotype stomatal patterning traits for QTL mapping in maize . bioRxiv 10.1101/2020.10.09.333880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Rosenow DT, Nguyen HT (2000) Stay green trait in grain sorghum: relationship between visual rating and leaf chlorophyll concentration. Plant Breed 119: 365–367 [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV (2010) The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SSD1. Plant Cell 22: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong Y-Y, Wang Y, Xu P, Ke S-D, Liu H-Y, Zhu J-K, Oliver DJ, Xiang C-B (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Peng S, Li Y (2019) Increase rate of light-induced stomatal conductance is related to stomatal size in the genus Oryza. J Exp Bot 70: 5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Park J-H, Lee S, Lee JH, Hwang D, Kwak JM, Kim YJ (2020) Regulation of stomatal development by stomatal lineage miRNAs. Proc Natl Acad Sci USA 117: 6237–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.